GEM-X 3′ v4 is the latest chemistry for the Chromium X system. It improves single-cell RNA sequencing by delivering faster partitioning (~6 minutes), higher sensitivity, greater throughput, and cleaner data compared with previous versions.

Single-cell RNA sequencing (scRNA-seq) has transformed our ability to study complex tissues, tumor heterogeneity, and immune microenvironments. By measuring gene expression at cellular resolution, scRNA-seq reveals rare populations and subtle transcriptional states invisible to bulk methods.
Yet as the technology matured, researchers faced recurring challenges:
Fragile cells, such as neutrophils, were often underrepresented.
Data sensitivity was limited by detection efficiency.
Throughput bottlenecks restricted large-scale studies.
Multiplets complicated downstream analysis.
To address these pain points, GEM-X 3′ v4 chemistry was developed for the 10x Genomics Chromium X series. This upgrade offers faster run times, improved sensitivity, higher throughput, and lower multiplet rates—collectively enhancing data quality for a wide range of applications.
One of the most striking improvements is the reduction in partitioning time. Previous chemistries required around 18 minutes for GEM (Gel Bead-in-Emulsion) formation. With GEM-X 3′ v4, that process now completes in ~6 minutes.
This shorter runtime reduces mechanical stress on fragile cells such as neutrophils and granulocytes. In pilot comparisons, GEM-X consistently captured more neutrophils while maintaining balanced proportions of other immune subsets. For researchers studying immunology, inflammation, or tumor-infiltrating immune cells, this improvement is particularly impactful.
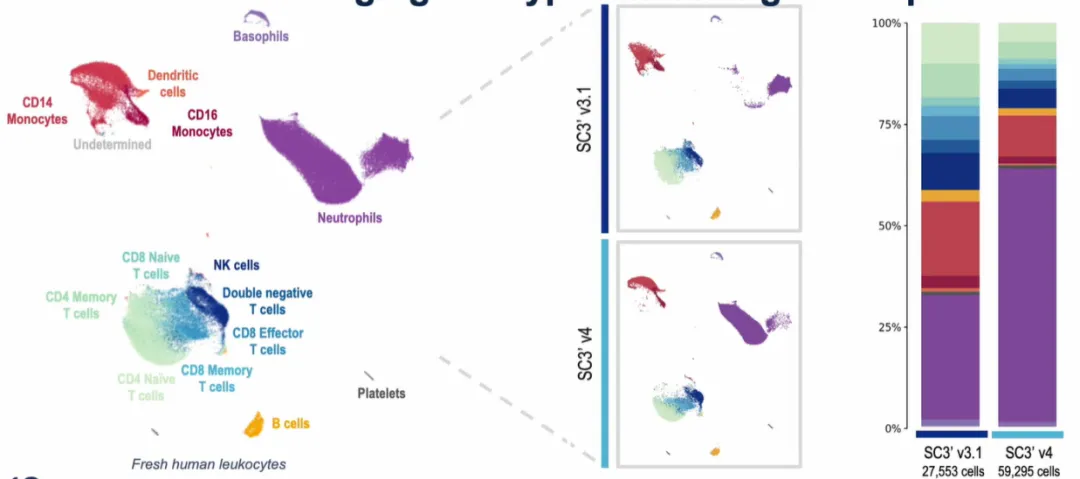
Figure 1. Neutrophil capture efficiency is significantly improved with GEM-X.
Data sensitivity—the number of unique genes and transcripts detected per cell—is a critical determinant of downstream analysis power.
Benchmarking studies have shown that GEM-X 3′ v4 can detect 50–100% more genes and up to ~2.5× more transcripts per cell compared to the 3′ v3.1 chemistry, at equivalent sequencing depth.
This enhanced sensitivity improves:
Low-abundance gene detection (e.g., transcription factors, cytokines).
Trajectory inference across subtle cell states.
Rare population analysis, where each transcript counts.
For researchers working on developmental biology or tumor heterogeneity, this gain in sensitivity translates directly into richer biological insights.
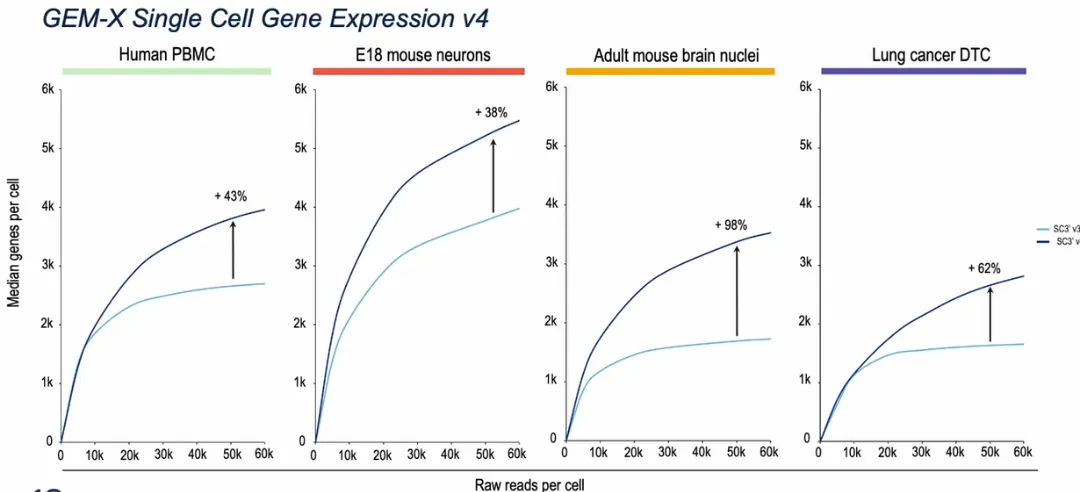
Figure 2. Comparison of median gene counts across different tissues between V3.1 and GEM-X (V4).
Another major advantage of GEM-X 3′ v4 is the increased throughput. Each channel on the Chromium X chip can now capture up to 20,000 cells, double the effective capacity of the previous generation
This boost offers several benefits:
Larger atlases per run, enabling more comprehensive tissue profiling.
Cost efficiency, as more cells are processed per sequencing lane.
Improved statistical power, especially for heterogeneous or rare populations.
For tumor microenvironment studies or organ-level atlases, higher throughput allows researchers to uncover rare cell states without needing multiple independent runs.
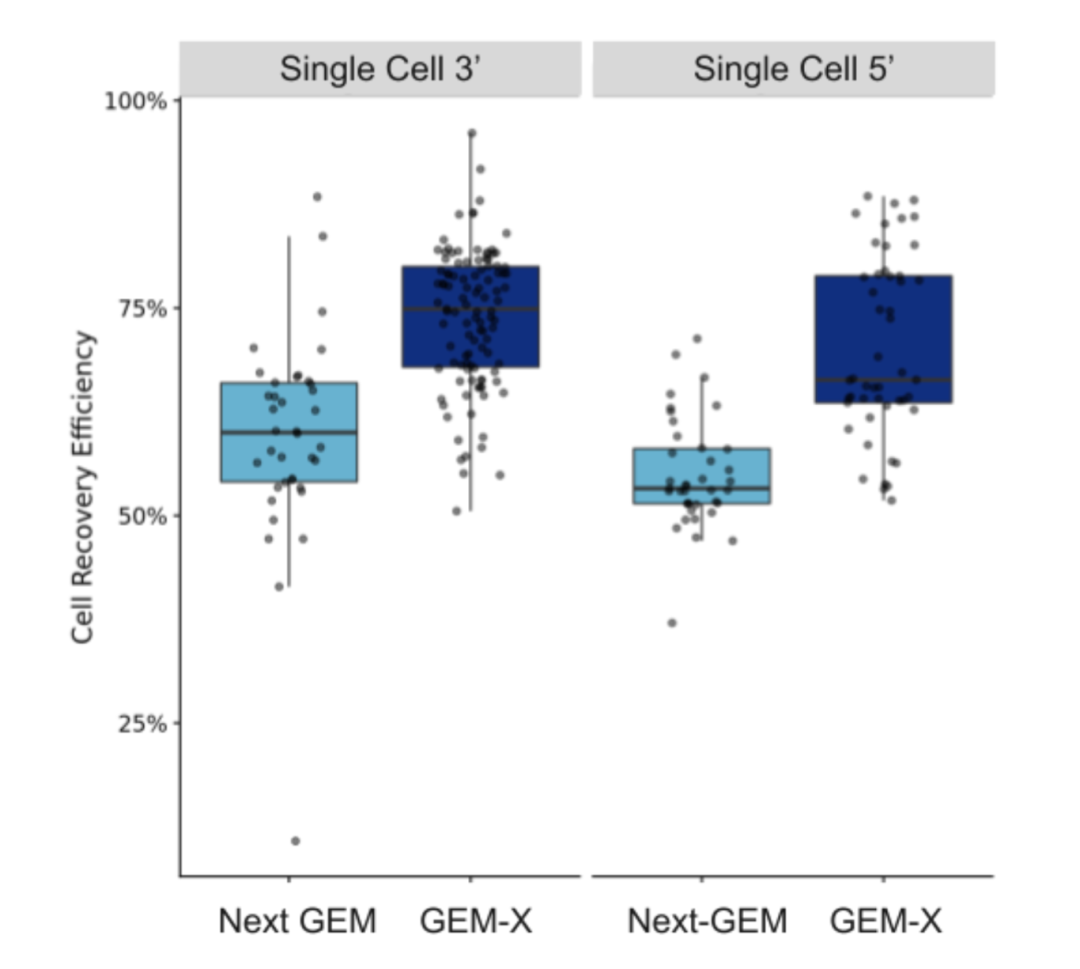
Figure 3. Comparison of cell capture efficiency between V3.1 and GEM-X (V4).
Multiplets (two or more cells encapsulated together) can complicate downstream analysis, leading to misassigned expression profiles. GEM-X 3′ v4 reduces multiplet rates by approximately 50% compared with v3.1, thanks to refined microfluidic designs and improved bead chemistry.
Lower multiplet rates mean:
Cleaner clustering with fewer ambiguous cells.
More accurate differential expression.
Less data discarded during QC filtering.
Together with higher sensitivity, these gains ensure datasets are both deeper and cleaner—qualities critical for high-stakes projects like biomarker discovery or clinical research.
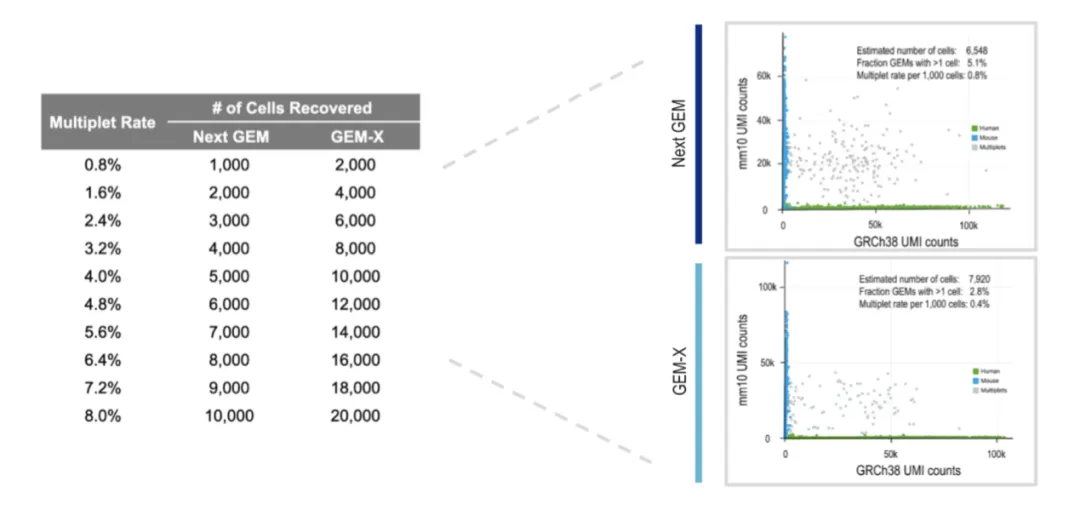
Figure 4. Comparison of multiplet rates between V3.1 and GEM-X (V4).
At Omics Empower, we provide end-to-end single-cell solutions powered by GEM-X 3′ v4 chemistry:
Whether your project involves cancer biology, immunology, developmental systems, or large-scale single-cell mapping, our GEM-X 3′ v4 service delivers faster, deeper, and cleaner datasets. Below are representative results from our in-house validation.
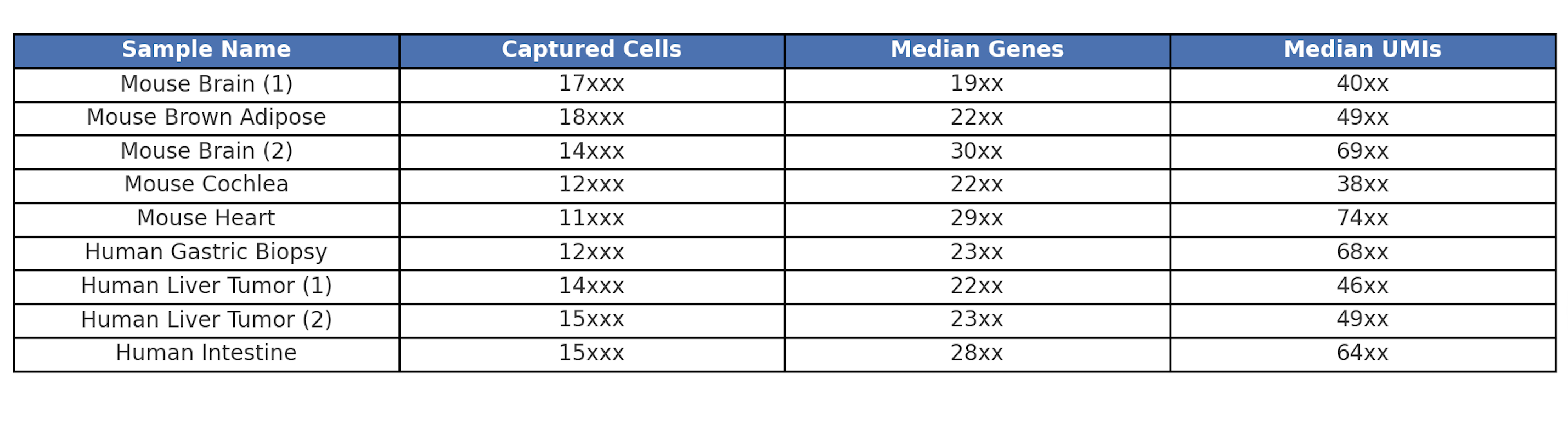
Figure 5. Omics Empower In-House Data - GEM-X 3' v4 Performance Across Samples
By addressing the limitations of earlier chemistries, GEM-X 3′ v4 establishes a new standard for single-cell RNA-seq. Faster GEM generation, higher sensitivity, expanded throughput, and lower multiplet rates collectively enhance data quality and unlock new biological insights.
At Omics Empower, we are proud to make this cutting-edge technology available to our collaborators and clients worldwide.
Contact us today to plan your GEM-X 3′ v4 single-cell RNA-seq project.
Credit: Figures labeled as “Omics Empower in-house data” are generated from our internal validation experiments using GEM-X 3′ v4 chemistry. Other figures are adapted from 10x Genomics official materials. 10x Genomics, Chromium, and GEM-X are trademarks of 10x Genomics, Inc. All rights reserved.
GEM-X 3′ v4 is the latest chemistry for the Chromium X system. It improves single-cell RNA sequencing by delivering faster partitioning (~6 minutes), higher sensitivity, greater throughput, and cleaner data compared with previous versions.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong