
Spatial transcriptomics is rapidly evolving, but can it truly reach single-cell resolution? With the release of Space Ranger v4.0, 10x Genomics has taken a critical step by integrating H&E-based cell segmentation into its Visium HD platform—enhancing biological accuracy while preserving spatial context.
Traditional spatial transcriptomics platforms preserve the spatial location of transcripts, but most do not directly capture expression at the single-cell level. Instead, data is collected in small capture areas (e.g., 2×2 μm bins in Visium HD), which are often aggregated to approximate a cell. However, cells are not perfect circles—they are irregular and heterogeneously distributed, especially in complex tissues. This approximation leads to loss of spatial fidelity.
To bridge this gap, cell segmentation aligns transcriptomic data with biologically accurate cell boundaries using histological images. This approach has already been adopted by other platforms, such as BGI’s Stereo-seq, which uses H&E or ssDNA staining to create cell-level data ("CellBins").
Now, 10x Genomics has joined the movement by integrating H&E-based cell segmentation directly into its Space Ranger v4.0 pipeline.
According to the latest technical documentation from 10x Genomics, Space Ranger v4.0 now fully supports H&E-based cell segmentation as part of its default analysis pipeline. Users can activate this function via a standard command:space ranger count.
The segmentation results are available in both the Web Summary report and Loupe Browser, making it easy to visualize and explore spatially resolved cell-level data.
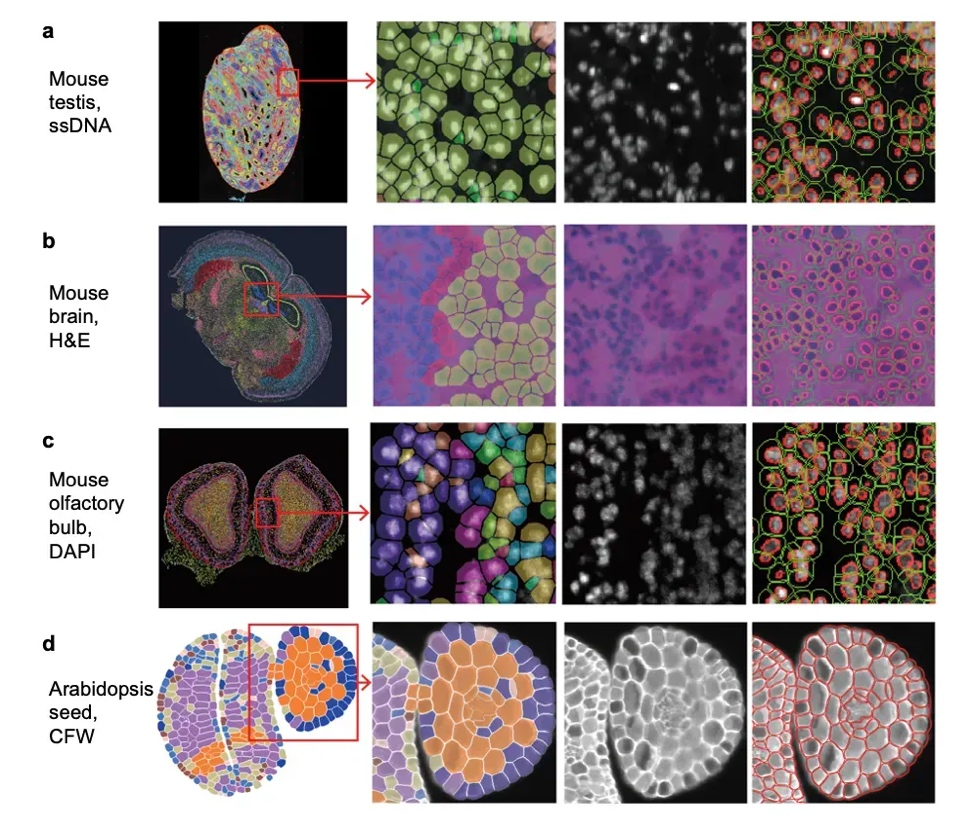
This segmentation module is built upon StarDist v6, a deep learning model designed to detect and segment star-convex nuclear structures—a common shape for cell nuclei in tissue.
The 10x Genomics team trained the model on an extensive dataset:
17,000+ image tiles
From over 150 H&E-stained images
Covering 24 tissue types across human and mouse
Including FFPE, fresh frozen (FF), and fixed frozen (FxF) samples
Tissue types used for training include:
Human tissues: Thymus, skin (melanoma), prostate, colon, colorectal cancer, breast, breast cancer, tonsil, brain, brain cancer, lung, lung cancer, and spleen.
Mouse tissues: Brain, bone, testis, small intestine, spleen, embryo, liver, lung, kidney, and thymus
The segmentation algorithm in Space Ranger follows a two-step process:
1. Nuclei Segmentation
StarDist detects nuclei from H&E images and outputs a nuclear mask file.
2. Cell Assignment
Spatial bins within a defined radius of each identified nucleus are grouped under a common cell ID.
This results in the generation of:
A new gene-barcode matrix
Updated visualization files (e.g., .cloupe)
Segmentation-aware data for downstream analysis
The demo dataset provided by 10x Genomics showcases segmentation results on a fresh frozen rhesus macaque kidney sample processed with Visium HD 3'. Segmentation output is visualized via downstream clustering and reveals clear cell-like contours reconstructed from bin aggregations.
While the boundaries appear jagged due to the bin-based nature of the data, the accuracy in nuclear localization and morphological fidelity remains high—making the method suitable for refined spatial analysis.
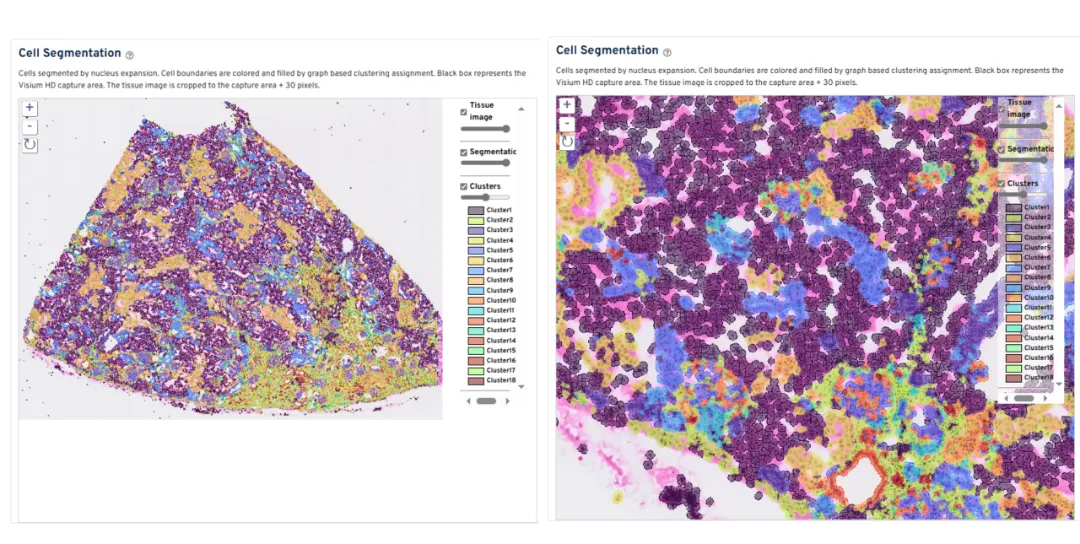
Image source: 10x Genomics official demo
The Web Summary report includes segmentation metrics, allowing users to evaluate performance and segmentation quality intuitively.
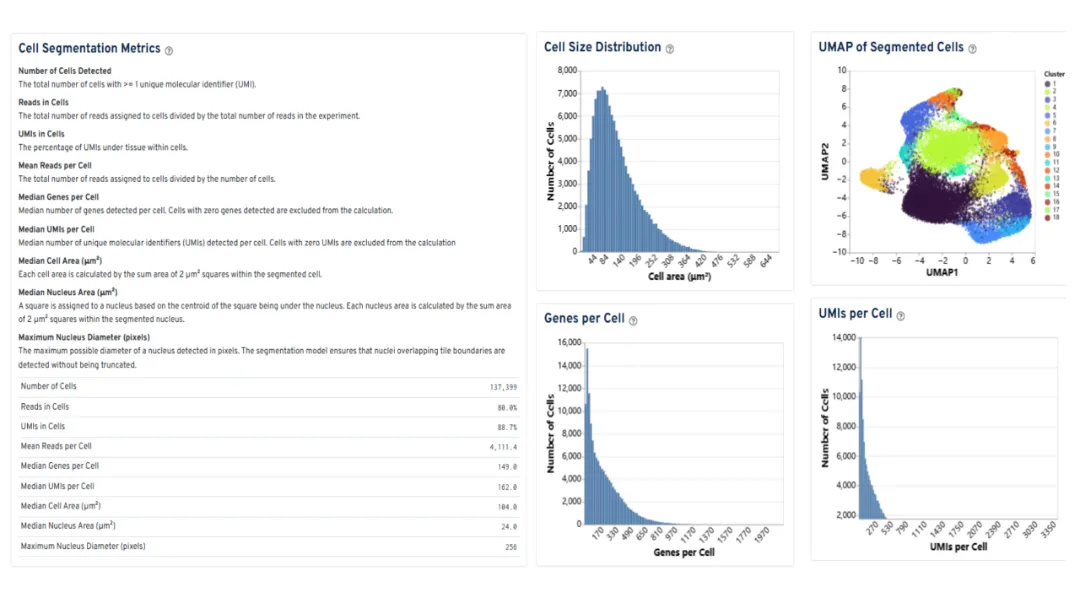
Image source: 10x Genomics official demo
While segmentation enhances resolution and mitigates bin-cell mismatches, it’s not a complete replacement for true single-cell technologies. Users should exercise caution in biologically complex tissues.
As stated in 10x Genomics' documentation, the segmentation model assumes approximately round nuclear morphology. This makes it less effective for cell types with irregular shapes, such as:
Neurons
Muscle cells
Plant cells
For such tissues, it is still advisable to:
Use bin-merging strategies
Or explore platforms like Xenium, which utilize multimodal imaging to achieve higher-fidelity segmentation across diverse morphologies
At Omics Empower, we have completed validation of Space Ranger v4.0 and are actively integrating its cell segmentation outputs into our spatial analysis workflows.
If you're encountering challenges in spatial transcriptomics, our expert technical team is here to help. Whether you're working with human or animal samples, we provide comprehensive, end-to-end support across the entire workflow, including:
Our spatial transcriptomics and single-cell sequencing services are tailored to diverse research areas such as oncology, neuroscience, and developmental biology. Reach out to explore how we can support your project from start to finish.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong