
Single-cell RNA sequencing (scRNA-seq) has revolutionized how we understand cellular behavior, from fate decisions and developmental transitions to responses under stress and disease. However, one step remains foundational to nearly all downstream analyses—cell type identification.
As many researchers will tell you, accurate cell type annotation is like laying a solid foundation for a house: if it's unstable, everything built on top becomes unreliable.
In this post, we walk you through common challenges in scRNA-seq cell type annotation—and how to tackle them with confidence.
Before jumping into marker-based annotation, take a moment to evaluate your clustering parameters—particularly the resolution. Resolution directly affects the number and separation of clusters, which in turn impacts your ability to distinguish meaningful cell types.
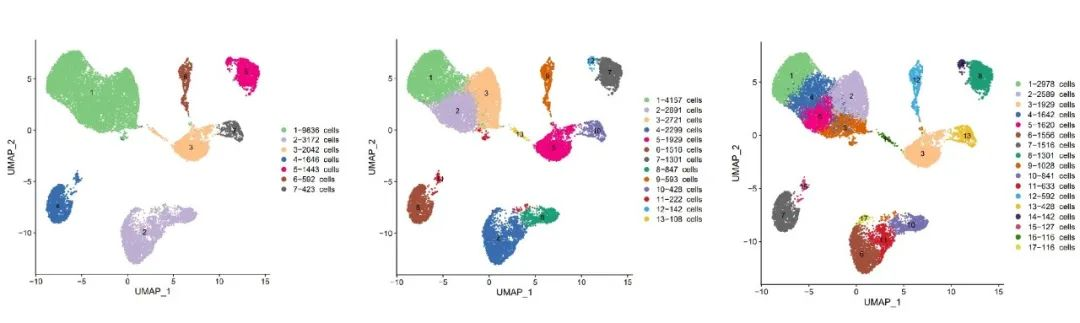
But does a higher resolution always lead to more identifiable cell types?
Not necessarily.
Key considerations:
Low resolution may merge distinct cell types into a single cluster, masking biological heterogeneity.
High resolution increases cluster count, but doesn’t necessarily reveal new cell types—just smaller subsets, complicating annotation.
Tips to optimize clustering:
1. Examine the top 10 markers per cluster—are they unique and specific?
2. Check for high inter-cluster similarity; if multiple clusters look alike, merging may be justified.
3. If one cluster shows markers from multiple cell types, try increasing the resolution.
Once clustering is optimized, it's time to annotate each cluster. There are two general approaches:
These tools match your data against curated datasets or marker gene libraries:
SingleR: Fast, easy-to-use (human and mouse only)
Celaref: Based on expression similarity between datasets
Garnett: Machine learning-based, uses marker rules
CellAssign: Bayesian framework using marker scores
These tools are excellent for preliminary annotation but may lack accuracy in complex or poorly characterized tissues.
This is still the gold standard—especially for high-impact publications.
How to manually annotate:
Search published literature for your tissue, organism, and disease model.
Compile marker gene lists from authoritative sources.
Use 2–3 well-established markers per cell type.
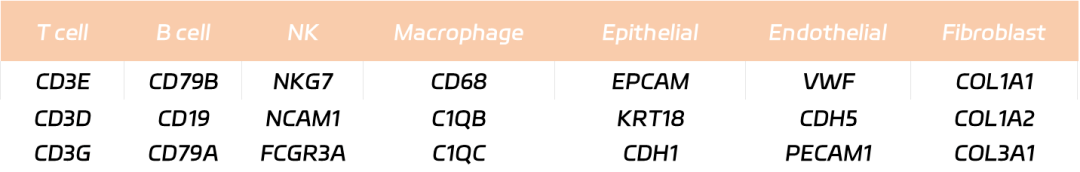
Useful databases:
Quickly explore UMAPs, feature plots, and expression levels without coding. Perfect for quality checks and presentations.
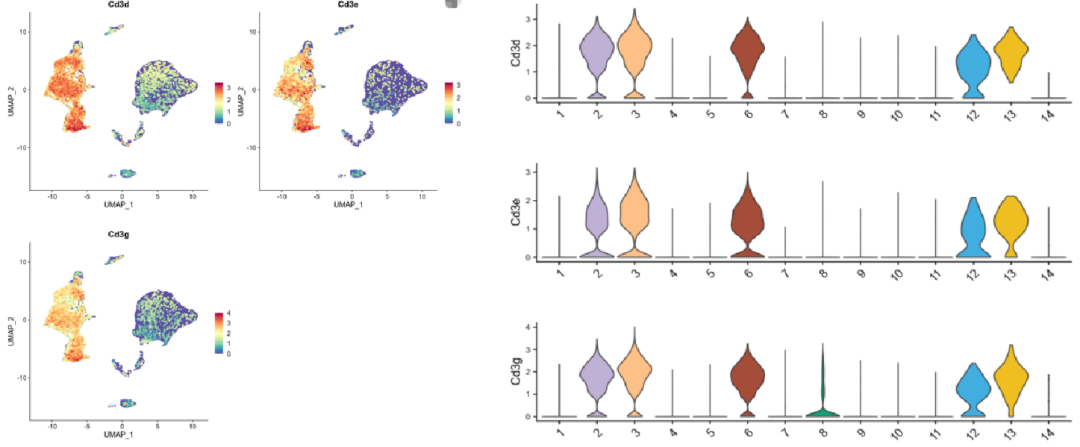
Use Seurat or similar tools to generate:
UMAP plots
Violin plots
Dot plots
Example: For T cell identification, examine expression of CD3D, CD3E, and CD3G. If clusters 2, 3, 6, 12, and 13 all express these markers, you can confidently label them as T cells.
Not every cluster will fit neatly into known cell types. Here’s how to troubleshoot:
Low-quality cells: Check for unusually low UMI counts.
Doublets: Co-expression of two unrelated cell-type markers.
Rare or novel populations: Use top markers and literature to infer identities.
Truly unknown: Label as "unknown" or "other" for now.
At Omics Empower, we specialize in end-to-end single-cell sequencing services tailored to your research needs. With over 350 publications and deep experience across human and animal models, our team ensures robust cell type annotation, insightful downstream analysis, and expert support every step of the way.

Need help annotating your single-cell data?
Get in touch to learn more about our customizable scRNA-seq services.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong