Yes—the published method and public browser target plants and reference plant atlases.

If you struggle to label plant cell types because classic markers don’t translate across species or tissues, the OMG Browser offers a no-code workflow based on Orthologous Marker Groups (OMGs). It maps your cluster markers to orthogroups, tests significance, and highlights likely cell-type matches—so you can move from clustering to biological interpretation faster. The approach was published in Nature Communications (Jan 2, 2025).
Unlike animal models, plant cell-type markers can be sparse or species-specific. Cross-species integration is also non-trivial and computationally heavy. The OMG framework side-steps this by comparing shared orthologous marker groups between your dataset and a reference atlas, then using Fisher’s exact test to quantify significance of overlap.
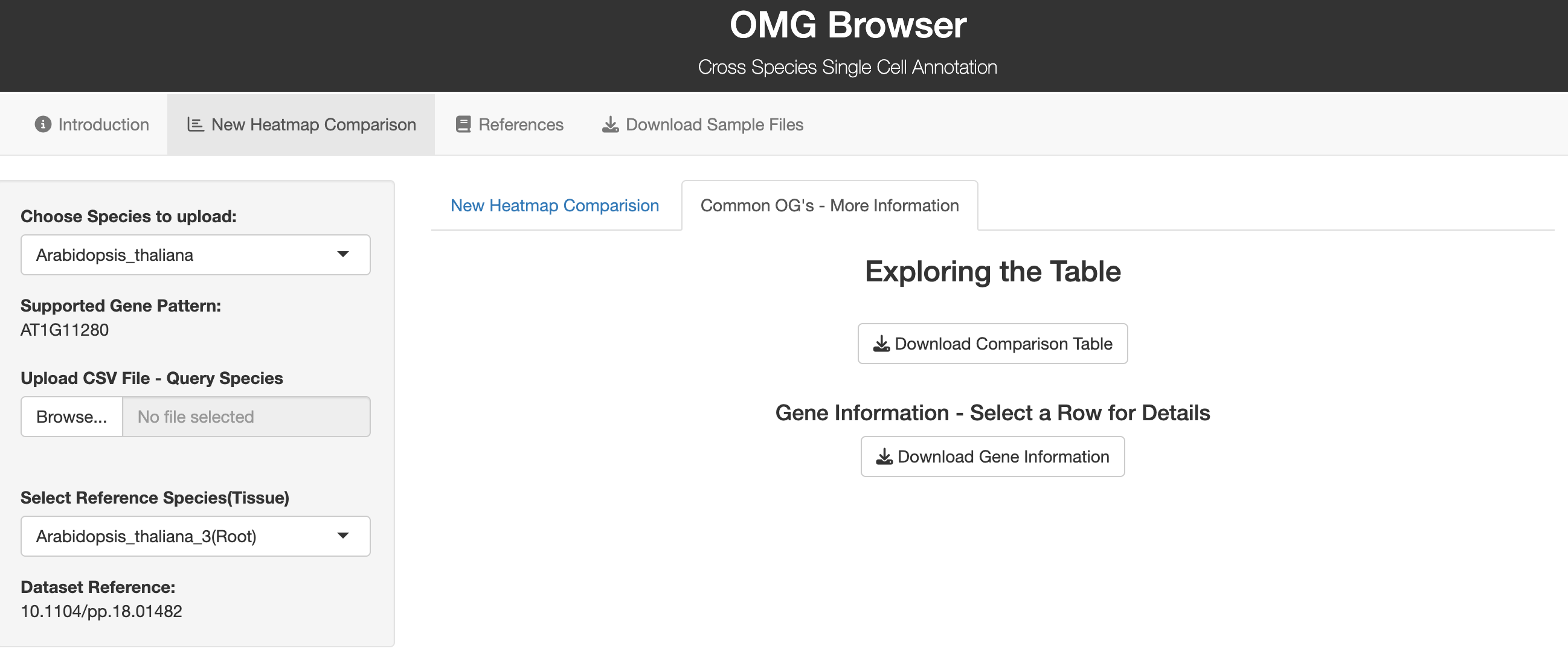
The OMG Browser is a web tool that implements the OMG workflow in a user-friendly way—no coding required. You upload cluster marker genes, select species and tissue, and get a heatmap showing −log10(p-value) for each cluster-to-cell-type comparison. Significant matches are boxed so you can quickly infer likely identities. The underlying method leverages OrthoFinder to build orthogroups and then performs statistics on shared markers.
1. Pick your target species. The browser shows accepted gene symbol formats for that species.
2. Upload your marker list (CSV). Make sure symbols match the required format (see the tool’s sample file).
3. Choose a reference species/tissue. The tool computes overlaps and renders a heatmap; download the results table for downstream QC.
Where to try it: OMG Browser (orthomarkergenes.org). The public GitHub repo documents the method and links to the live browser.
The public materials and repo list multiple plant species commonly used in the workflow (e.g., Arabidopsis, tomato, rice, maize, poplar, tobacco, cotton, periwinkle, etc.), with a broader 15-species atlas used in benchmarking. Always check the latest browser build for coverage before analysis.
Clustering granularity. Extremely fine clustering can produce small clusters with too few reliable markers, reducing annotation confidence. Start with a biologically reasonable resolution, then refine. (Discussed in the method.)
Marker selection. Use conservative thresholds (e.g., min % expressed, effect size) and consider combining methods to derive robust marker sets before upload.
Gene symbols. Ensure exact species-specific naming; mismatches will silently reduce overlap counts.
Species/tissue match. Choose the closest available tissue context in the reference to avoid spurious matches.
Interpretation ≠ ground truth. The output highlights significant overlaps; confirm with orthogonal evidence (in situ, known developmental trajectories, motif enrichment, etc.).
Omics Empower supports end-to-end plant single-cell sequencing projects—particularly where cell walls and RNases make dissociation and RNA integrity challenging.
We support academic labs, biotech companies, and research hospitals with reliable, full-process project execution—from sample preparation to bioinformatics analysis.
With labs across Asia, Europe, and North America, we offer localized project handling, quicker turnaround times, and responsive communication throughout every stage.
Reference
Chau, T.N., Timilsena, P.R., Bathala, S.P. et al. Orthologous marker groups reveal broad cell identity conservation across plant single-cell transcriptomes. Nat Commun 16, 201 (2025). https://doi.org/10.1038/s41467-024-55755-0
Yes—the published method and public browser target plants and reference plant atlases.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong