Mapping protein–protein interactions (PPIs) among membrane proteins is a long-standing challenge in molecular biology. Traditional GAL4-based Yeast Two-Hybrid (Y2H) systems rely on nuclear localization of fusion proteins, making them unsuitable for receptors, channels, and transporters that remain embedded within lipid bilayers. To enable interaction studies directly at the membrane, the Membrane Yeast Two-Hybrid (MbY2H) system was developed.
Its success, however, depends critically on selecting the correct bait and prey vectors so that the split-ubiquitin fragments—NubG and Cub-LexA-VP16—are correctly oriented on the cytosolic side of the membrane.
Understanding these requirements is central to generating reliable MbY2H results.
What Is the Membrane Yeast Two-Hybrid (MbY2H) System?
The Need for a Membrane-Compatible Yeast Two-Hybrid System
Classical GAL4-based Y2H assays require both bait and prey fusion proteins to enter the nucleus, where GAL4 DNA-binding and activation domains reconstitute transcriptional activity. This makes GAL4-Y2H unsuitable for:
Integral membrane proteins
Multi-pass membrane transporters
Receptors with luminal or extracellular domains
Proteins that must remain at the plasma membrane or endomembrane system
To overcome these constraints, researchers developed the split-ubiquitin Yeast Two-Hybrid system, now widely known as the Membrane Yeast Two-Hybrid (MbY2H) system.
How the Split-Ubiquitin Y2H System Works
The split-ubiquitin system differs fundamentally from classical Y2H:
NubG (mutated N-terminal half) and Cub (C-terminal half fused to LexA-VP16).
Bait proteins are fused to Cub-LexA-VP16 and anchored in the membrane.
Prey proteins are fused to NubG, either at the N-terminus (NubG-X) or C-terminus (X-NubG).
When bait and prey interact on the cytosolic side of the membrane, NubG and Cub reconstitute ubiquitin.
Yeast ubiquitin-specific proteases (UBPs) recognize the restored ubiquitin and cleave Cub, releasing LexA-VP16.
LexA-VP16 translocates to the nucleus and activates LexA reporters (such as HIS3, ADE2, or LacZ).
Why MbY2H Is Essential for Y2H Membrane Screening
The Membrane Yeast Two-Hybrid system enables PPIs to be studied:
Without forcing membrane proteins into the nucleus
With native orientation of N- and C-termini preserved
At the membrane, where many interactions naturally occur
For anyone targeting keywords such as Yeast Two-Hybrid, split ubiquitin Y2H, membrane protein Y2H, or Y2H screening, the MbY2H system represents the modern evolution of the classical Y2H assay—optimized specifically for membrane biology.
For more background on the evolution from GAL4 to split-ubiquitin systems, see our related article: Yeast Two-Hybrid (Y2H) Systems: From GAL4 to Split-Ubiquitin Assays
Choosing the Right MbY2H Bait Vector (pBT3 Series)
pBT3-N: For N-Cytosolic / C-Luminal Proteins
If your bait protein has an N-terminus facing the cytosol and a C-terminus oriented toward the luminal or extracellular space, pBT3-N provides the correct orientation for Cub-LexA-VP16.
This vector is suitable for Type II membrane proteins and many single-pass proteins whose cytosolic tails mediate interactions.
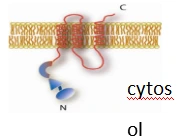
Figure 1. Representative topology of a Type II membrane protein used in pBT3-N bait construction.
pBT3-SUC: For Luminal N-Termini Containing Signal Peptides
Some membrane proteins begin with a signal peptide that directs them into the secretory pathway. However, many mammalian signal peptides do not function efficiently in yeast.
In such cases, pBT3-SUC, which includes the yeast SUC2 signal sequence, ensures proper membrane targeting.
It is ideal for proteins with luminal or extracellular N-termini and cytosolic C-termini that contain signal peptides.
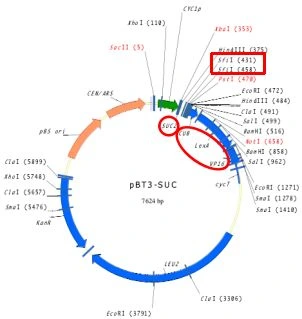
Figure 2. Map of the pBT3-SUC vector showing the SUC2 signal sequence upstream of the cDNA insertion site.
The diagram illustrates that the SUC2 signal peptide is positioned upstream of the cDNA cloning site (SfiI), while the Cub–LexA–VP16 fusion is located downstream. Upon expression, the resulting bait fusion protein contains SUC2 at the N-terminus and Cub–LexA–VP16 at the C-terminus. This architecture ensures correct membrane targeting in yeast and positions Cub on the cytosolic side, allowing efficient interaction with NubG-tagged prey proteins in the split-ubiquitin MbY2H system.
pBT3-STE: For Luminal N-Termini Without Signal Peptides
Some proteins lack a classical signal peptide but still orient with a luminal N-terminus via internal signal-anchor sequences.
pBT3-STE accommodates these noncanonical membrane insertion patterns, ensuring Cub-LexA-VP16 remains cytosolic.
Proteins with Both Termini Facing the Cytosol
If both termini are cytosolic, either pBT3-N or pBT3-STE can be used.
Researchers often compare both constructs to determine which produces better expression and lower background.
Choosing the Right MbY2H Prey Vector (pPR3 Series)
MbY2H is not only defined by bait topology; prey orientation also determines whether NubG is accessible to Cub-LexA-VP16 on the cytosolic side. The system uses two prey vector formats:
NubG-X (pPR3-N)
X-NubG (pPR3-C)
These two libraries differ in how NubG is fused to the prey protein.
pPR3-N (NubG-X): High-Complexity Default Library
NubG fused to the N-terminus of prey proteins functions well for most applications.
This library features high complexity (≈10⁷ clones) and captures large cDNA inserts (>1.5 kb), making it ideal for broad interactome screens.
pPR3-C (X-NubG): Required for Type I Membrane Preys
For Type I membrane proteins—with luminal N-termini and cytosolic C-termini—NubG must be placed at the C-terminus to ensure cytosolic accessibility.
pPR3-C provides this orientation.
Still Unsure Which Vector to Use? We Can Help
If you are planning a Yeast Two-Hybrid or Membrane Yeast Two-Hybrid study and would like assistance with vector selection, library construction, or screening design, our team can help.
At Omics Empower, we provide integrated yeast hybrid solutions covering Yeast One-Hybrid (Y1H), Yeast Two-Hybrid (Y2H), Membrane Yeast Two-Hybrid (MbY2H), and custom cDNA library construction and screening. Our platforms support systematic investigation of both DNA–protein and protein–protein interactions, enabling researchers to explore gene regulation, signaling pathways, and molecular networks with confidence.
Why Choose Omics Empower yeast two-hybrid (Y2H) Service?
As one of the earliest groups in Asia to commercialize yeast hybrid technologies, we have more than a decade of experience in Y1H and Y2H assay development. Our team has mastered both Gateway and SMART library construction workflows and applies multiple yeast hybrid systems—including classical GAL4-based Y2H and split-ubiquitin assays—to meet diverse experimental needs.
Our yeast library platform has supported 200+ customer publications, including studies featured on the covers of Science and Cell, demonstrating the reliability and scientific impact of our data. With rigorous quality control, high-efficiency transformation, and dedicated technical support, we help ensure that each Y2H or MbY2H project begins with robust experimental design and ends with reproducible, publication-ready results.
If you would like to discuss your Y1H, Y2H, or MbY2H project, our scientific team is ready to assist with vector selection, library strategy, and screening workflows.







_Screening.webp)