Single-cell transcriptomics and spatial transcriptomics have rapidly advanced plant biology by enabling high-resolution analysis of gene expression at both cellular and tissue levels. Together, these technologies provide a powerful framework for resolving cellular heterogeneity, tissue organization, and developmental dynamics in plants.
By integrating single-cell RNA sequencing (scRNA-seq) with spatial transcriptomics, transcriptional states can be directly linked to their spatial context, allowing the construction of detailed molecular maps of plant tissues. This combined approach is increasingly used to study cell differentiation, tissue patterning, and plant responses to environmental cues.
Despite these advantages, applying single-cell and spatial transcriptomics to plant samples remains technically demanding. Plant-specific structural and biochemical features introduce challenges that complicate both experimental workflows and data interpretation. Here, we summarize key challenges, methodological strategies, and practical considerations for plant single-cell and spatial transcriptomics studies.
Key Challenges in Plant Single-Cell and Spatial Transcriptomics
1. Cell Wall Barriers in Single-Cell Isolation
A fundamental challenge in plant single-cell transcriptomics is the presence of rigid cell walls. Generating viable protoplasts requires enzymatic digestion of the cell wall, which is highly sensitive to species, tissue type, and developmental stage.
As a result:
Protocols are difficult to standardize across plant species
Experimental reproducibility may vary significantly
Workflow optimization often becomes highly sample-specific
These factors make protoplast-based scRNA-seq in plants substantially more complex than in animal systems.
2. Dissociation-Induced Stress and Technical Bias
Cell dissociation can trigger stress responses in plant cells, leading to the activation of defense-related genes and the introduction of transcriptional artifacts. Additional technical biases include:
To mitigate these issues, single-nucleus RNA sequencing (snRNA-seq) has gained popularity as an alternative approach. Nuclei isolation can reduce dissociation-induced artifacts, although it may result in partial loss of cytoplasmic transcripts.
3. Structural Complexity in Spatial Transcriptomics
Spatial transcriptomics introduces additional technical challenges due to the structural diversity of plant tissues. Factors affecting cryosectioning and tissue integrity include:
These characteristics increase the risk of tissue fragmentation, detachment from slides, and uneven RNA capture, thereby complicating spatial transcriptomics experiments.
4. Platform Adaptation for Plant Samples
Spatial transcriptomics platforms have advanced rapidly in recent years. For instance, 10x Genomics has improved spatial resolution from 100 μm to 2 μm.
However, current high-resolution configurations are primarily optimized for mammalian tissues. For plant samples, spatial resolution and protocol compatibility remain areas requiring further optimization and methodological innovation.
5. Data Analysis Challenges in Non-Model Species
From a computational perspective, plant single-cell and spatial transcriptomics studies face several obstacles:
Limited availability of validated marker genes
Incomplete genome annotations
Underdeveloped reference databases for non-model species
These limitations complicate cell-type annotation, functional interpretation, and cross-study comparisons, particularly in non-model plant systems.
Methodological Strategies for Plant Single-Cell and Spatial Transcriptomics
Single-Cell and Single-Nucleus Transcriptomics Using 10x Chromium
The 10x Chromium platform integrates microfluidics and barcoding technologies to enable high-throughput profiling of individual cells or nuclei.
In plant research, both protoplast-based scRNA-seq and nuclei-based snRNA-seq can be applied depending on tissue characteristics and experimental objectives. These approaches support:
Identification of cell types and states
Analysis of cellular heterogeneity
Reconstruction of developmental trajectories
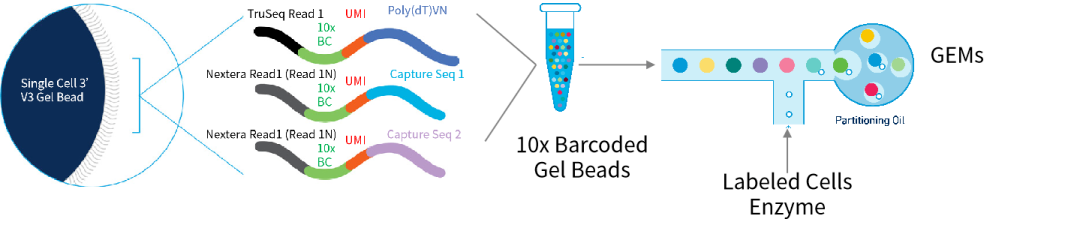
Figure 1. Structure of 10x Chromium gel beads and formation of gel bead–in–emulsions (GEMs). Source: 10x Genomics.
Spatial Transcriptomics Using 10x Visium
The 10x Visium Spatial Gene Expression platform incorporates spatial barcoding to preserve positional information while quantifying gene expression.
By integrating spatial coordinates with transcriptomic data, researchers can:
Map gene expression patterns across tissue sections
Identify spatially localized cell populations
Investigate tissue organization and developmental gradients
Spatial transcriptomics is particularly valuable for understanding complex plant tissue architecture and functional compartmentalization.
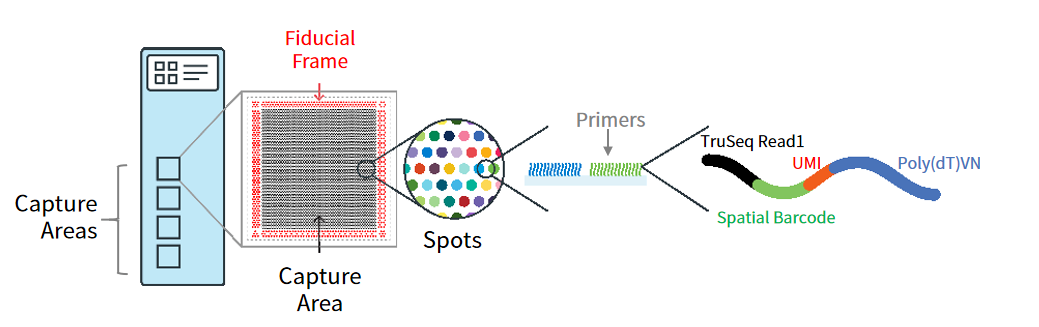
Figure 2. Schematic overview of the capture area architecture of the 10x Visium spatial transcriptomics slide. Source: 10x Genomics.
Our Case Study: Integrating Single-Cell and Spatial Transcriptomics in Tree Development
An illustrative example of plant spatial transcriptomics integration was reported in Plant Communications (2023), where single-cell RNA sequencing and spatial transcriptomics were combined to investigate cambium differentiation during primary and secondary growth in poplar.
In this study, Omics Empower provided single-cell RNA sequencing and spatial transcriptomics services based on 10x Genomics scRNA-seq and Visium.
Through the integration of single-cell and spatial data, the study:
Identified distinct cell populations across primary and secondary growth tissues
Reconstructed differentiation trajectories of cambial cells
Revealed gene regulatory networks underlying xylem and phloem precursor development
This work demonstrates how coordinated experimental execution and integrative analysis of single-cell and spatial transcriptomics can resolve dynamic molecular processes in woody plant development that are difficult to capture using bulk or tissue-level approaches alone.
Practical Considerations for Sample Preparation
Single-Cell / Single-Nucleus Transcriptomics
Whole plants can be transported for protoplast-based workflows if properly stabilized
For nuclei-based workflows, fresh, low-starch tissues are preferred
Rapid freezing in liquid nitrogen and storage at −80 °C or shipment on dry ice is recommended
Spatial Transcriptomics
Target tissues should be trimmed to appropriate size prior to embedding
OCT embedding should fully infiltrate tissue structures
Vacuum infiltration can improve embedding quality in high-moisture or loosely connected tissues
Rapid freezing on dry ice followed by −80 °C storage is recommended
Need Support for Plant Single-Cell and Spatial Transcriptomics?
Plant single-cell and spatial transcriptomics projects often involve plant-specific challenges, from sample preparation and tissue handling to data interpretation across complex structures and non-model species.
Since 2018, Omics Empower has been providing single-cell transcriptomics services and has built extensive experience supporting plant-focused single-cell and spatial transcriptomics research. In 2020, we supported a plant single-cell RNA sequencing study on Arabidopsis thaliana published in Molecular Plant. At the time, this work was among the relatively few plant single-cell transcriptomics studies enabled through collaboration with a research service provider, reflecting early efforts to adapt single-cell technologies to plant systems.
If you are planning a plant single-cell or spatial transcriptomics study—or troubleshooting challenging plant samples—we welcome technical discussions to help define an appropriate workflow and analysis plan.
Reference
Li, R., Wang, Z., Wang, J.-W., & Li, L. (2023). Combining single-cell RNA sequencing with spatial transcriptome analysis reveals dynamic molecular maps of cambium differentiation in the primary and secondary growth of trees. Plant Communications, 4(5), 100665. https://doi.org/10.1016/j.xplc.2023.100665