Since the 19th-century formulation of cell theory, biologists have sought to decode how individual cells collectively shape the complexity of life. Every organ, tissue, and physiological function originates from the intricate interplay of diverse cell types. To understand life’s mechanisms, scientists must first understand the cell.
Single-cell sequencing has transformed this pursuit. By capturing molecular information from thousands of cells simultaneously, it allows researchers to uncover how distinct cell populations contribute to development, disease, and regeneration. Yet, despite its power, conventional single-cell RNA sequencing (scRNA-seq) presents technical barriers that can distort biological interpretation.
The emergence of single-nucleus RNA sequencing (snRNA-seq) has addressed these challenges by focusing on the nucleus—the most stable and information-rich compartment of the cell. This method now enables robust transcriptomic profiling from both fresh and frozen tissues, extending single-cell analysis to previously inaccessible specimens.

Limitations of Conventional scRNA-seq
Although scRNA-seq has been the cornerstone of transcriptomics for over a decade, its reliance on live, intact cells introduces several major constraints:
1. Reliance on Fresh Samples
scRNA-seq requires viable, intact cells, making it unsuitable for frozen or archived tissues. Many clinical and biobank specimens are cryopreserved to preserve RNA integrity, but once cells lose viability, they can no longer be processed for scRNA-seq. This restriction limits the use of valuable patient-derived materials and reduces experimental throughput.
2. Dissociation-Induced Artifacts
Tissue dissociation—often performed at 37 °C with proteases—can artificially induce stress-response genes, altering transcriptional profiles and introducing bias. van den Brink et al. demonstrated that dissociation triggers stress pathways, skewing cell-type identification [1]. Similar findings from Adam et al. showed that standard enzymatic digestion causes artifactual transcriptomic changes [2]. Denisenko et al. later confirmed that low-temperature processing significantly reduces these artifacts [3].
3. Incomplete Cell Recovery
Some cell types are lost during enzymatic digestion, particularly in solid organs such as brain, heart, and kidney. Fragile or deeply embedded cells may fragment or remain uncollected, leading to biased datasets. For example, dentate gyrus neurons and glomerular podocytes are under-represented in scRNA-seq results [4-6]. Filtration steps that exclude large or irregular cells further distort cell-type composition, undermining the accuracy of downstream biological interpretations.
The Rise of Single-Nucleus RNA Sequencing (snRNA-seq)
snRNA-seq isolates nuclei rather than intact cells, eliminating the need for live-cell dissociation. The method traces back to early genomic analyses by Navin et al., who profiled copy-number variation from single tumor nuclei [7]. Grindberg et al. later pioneered RNA sequencing from isolated nuclei of post-mortem brain tissue [8].
Building on these breakthroughs, subsequent methods such as Div-seq, DroNc-seq, and Cumulus advanced throughput and automation. Research teams led by Aviv Regev and Kun Zhang demonstrated that snRNA-seq can produce high-quality transcriptomic data from frozen clinical and brain samples [9-11]. These studies established snRNA-seq as a robust alternative to scRNA-seq, capable of revealing cell-type diversity and dynamic transcriptional states.
Key Advantages of snRNA-seq
1. Broader Sample Compatibility
The nuclear membrane remains intact even when cell membranes rupture, enabling snRNA-seq to work seamlessly with frozen, FFPE, and archived specimens. This has opened access to valuable biobank and clinical materials. Because the workflow relies on mechanical disruption rather than enzymatic digestion, it is simpler, more stable, and highly reproducible [3, 8, 9].
2. Accurate Transcriptional Representation
Frozen tissues preserve RNA at the moment of collection. With transcription halted, snRNA-seq captures unbiased, “snapshot-level” expression profiles, free from dissociation-induced stress. Studies show strong correlation between nuclear and cytoplasmic RNA data, confirming that snRNA-seq faithfully represents genuine transcriptional states [3-5].
3. Comprehensive Cell-Type Coverage
snRNA-seq recovers nuclei from all cell populations—including those that are fragile or hard to dissociate—yielding a more complete cellular atlas. This advantage is consistent across tissues such as brain, kidney, heart, and pancreas [5-9]. Researchers can now detect rare subpopulations and previously missing developmental or disease-specific states.
4. Stability and Reproducibility
snRNA-seq demonstrates excellent reproducibility between biological replicates. Consistent clustering patterns across independent samples confirm its reliability for multi-center studies and long-term data integration [10, 11].
5. Insight into Dynamic and Pathological States
Despite focusing on nuclear transcripts, snRNA-seq effectively captures transcriptional dynamics such as neuronal activation [12], cardiac regeneration [13], renal fibrosis [14], and developmental trajectories in heart and pancreas [15, 16]. The method reveals disease-associated expression changes comparable to or clearer than those identified by scRNA-seq.
snRNA-seq Is Becoming the Mainstream Approach
Publications using snRNA-seq have surged dramatically in recent years. Leading research institutions increasingly prefer nuclear over cellular sequencing for complex or clinically derived tissues. The trend reflects not only methodological reliability but also compatibility with emerging multi-omics strategies, where chromatin accessibility and transcriptomic data are integrated within the same nucleus.
Computational frameworks such as Cumulus and Seurat v5 now support efficient analysis of large-scale nuclear datasets. Looking forward, snRNA-seq will underpin high-throughput and multi-modal studies essential to building complete cellular atlases.
10x Genomics-Based snRNA-seq: The Industry Standard
Among commercial platforms, 10x Genomics has become the benchmark for single-nucleus sequencing. Its Chromium system supports both 3′ and 5′ gene-expression assays and delivers consistent, high-quality data across tissue types. Comparative evaluations have confirmed 10x Genomics as the most stable and accurate solution among leading technologies [11, 17].
Key benefits of 10x Genomics snRNA-seq include:
High capture efficiency (> 100 k nuclei per run)
Compatibility with fresh, frozen, and FFPE samples
Integration with single-cell and spatial transcriptomic workflows
Because of its reproducibility and flexibility, 10x Genomics snRNA-seq has become the preferred platform for human cell-atlas projects and translational research worldwide.
About Omics Empower snRNA-seq Service
Omics Empower is one of the earliest service providers to offer comprehensive single-cell and single-nucleus sequencing across Asia, Europe, and North America. Equipped with 10x Genomics and Mobidrop platforms, we deliver end-to-end solutions for fresh, frozen, and FFPE samples.
Here’s an example of our experimental data.

Figure 1. Trypan blue staining of isolated nuclei
Microscopic examination of the prepared nuclei after trypan blue staining shows that cell debris and impurities account for less than 10%. The nuclei concentration exceeds 1,000 nuclei/μL, with a total yield greater than 100,000 nuclei—meeting the quality requirements for 10x Genomics sequencing.
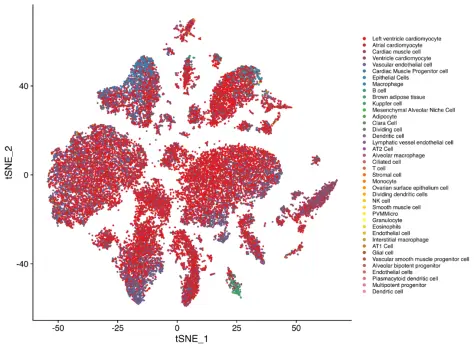
Figure 2. Cell-type identification
Single-nucleus RNA sequencing (snRNA-seq) of mouse heart tissue successfully identified all major cardiac cell types based on key marker genes, demonstrating comprehensive coverage of cardiomyocytes, endothelial cells, fibroblasts, and other supporting cell populations.
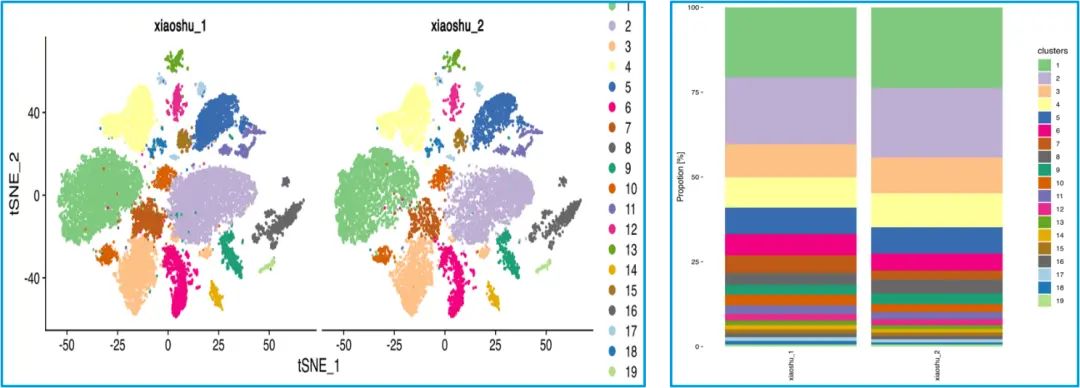
Figure 3. Technical reproducibility assessment
snRNA-seq analysis of mouse heart tissue demonstrates high reproducibility between two biological replicates, with consistent clustering patterns and comparable transcriptomic profiles across samples.
By integrating snRNA-seq with our single-cell and spatial transcriptomics platforms, Omics Empower helps researchers obtain accurate, bias-free transcriptomic maps—even from archived clinical materials—empowering discoveries in developmental biology, oncology, and regenerative medicine.
Why Researchers Choose Omics Empower
Proven scientific impact: Our clients have published 400+ peer-reviewed papers using data generated from Omics Empower’s single-cell and spatial transcriptomics services, including articles in Nature, Science, and Cell journals.
Expertise in complex samples: We have optimized nuclei isolation and QC pipelines for challenging tissues such as brain, heart, kidney, and tumors—achieving high nucleus purity and yield for 10x Genomics library preparation.
Reliable and reproducible data: Each project is processed under strict quality standards to ensure high sensitivity, minimal doublets, and accurate cell-type annotation.
End-to-end bioinformatics: From raw data processing to clustering, trajectory analysis, and publication-ready visualization, our in-house bioinformatics team delivers complete and interpretable results.
Integrated multi-omics capability: Omics Empower combines snRNA-seq with single-cell RNA-seq, ATAC-seq, and spatial transcriptomics to provide a multi-dimensional understanding of cellular states.
References
[1] van den Brink SC et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017; 14(10): 935-936.
[2] Adam M et al. Psychrophilic proteases dramatically reduce single-cell RNA-seq artifacts: a molecular atlas of kidney development. Development. 2017; 144(19): 3625-3632.
[3] Denisenko E et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020; 21(1): 130.
[4] Grindberg RV et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci USA. 2013; 110(49): 19802-19807.
[5] Wu H et al. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019; 30(1): 23-32.
[6] Koenitzer JR et al. Single-nucleus RNA-seq profiling of mouse lung: reduced dissociation bias and improved detection of rare cell types compared with single-cell RNA-seq. bioRxiv. 2020; doi: 10.1101/2020.03.06.981407.
[7] Navin N et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011; 472(7341): 90-94.
[8] Habib N et al. Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016; 353(6302): 925-928.
[9] Lake BB et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016; 352(6293): 1586-1590.
[10] Bakken TE et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018; 13(12): e0209648.
[11] Slyper M et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 2020; 26(5): 792-802.
[12] Lacar B et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016; 7: 11022.
[13] Hu P et al. Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts. Genes Dev. 2018; 32(19-20): 1344-1357.
[14] Lake BB et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat Commun. 2019; 10(1): 2832.
[15] Cui M et al. Dynamic transcriptional response to injury of regenerative and non-regenerative cardiomyocytes revealed by single-nucleus RNA sequencing. Dev Cell. 2020; 53(1): 102-116.
[16] Tosti L et al. Single-nucleus and in situ RNA sequencing reveals cell topographies in the human pancreas. bioRxiv. 2019; doi: 10.1101/733964.
[17] Ding J et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol. 2020; 38(6): 737-746.