Flow cytometry is a powerful technique for characterizing individual cells by measuring their physical and molecular features as they pass through a laser-based detection system. By labeling cells with fluorescent antibodies or dyes, researchers can rapidly analyze heterogeneous populations and quantify marker expression at single-cell resolution.
Fluorescence-activated cell sorting (FACS) builds on this concept by not only analyzing cells but also physically separating defined populations into collection tubes using electrostatic deflection. This makes FACS an essential tool for preparing high-quality samples for single-cell sequencing, where cell purity and viability directly influence downstream data quality.
Because we routinely support researchers in preparing and sorting samples for single-cell projects, we have gathered practical, experience-based guidance that reflects the questions scientists most often ask when using FACS for sequencing.
How do you choose the right nozzle size for flow cytometry sorting?
Nozzle size determines sorting pressure, droplet stability, and the level of mechanical stress applied to cells. Most instruments offer 70-µm, 100-µm, and 130-µm nozzle options, each with different impacts on viability.
A practical rule is to select a nozzle that is four to six times the diameter of the target cells. Most PBMCs and many tissue-dissociated cells sort well using a 100-µm nozzle (Figure 1). Larger or fragile cells—such as hepatocytes or neurons—typically perform better with a 130-µm nozzle due to lower pressure and gentler handling.
Because single-cell sequencing requires high viability, choosing a larger nozzle is often the safer option even if the sorting process becomes slightly slower.
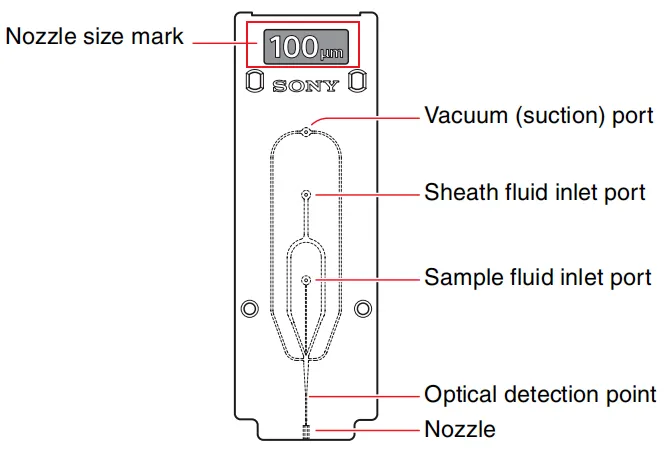
Figure 1. Flow cytometry nozzle structure and components.
Does flow cytometry sorting impose limitations on certain cell types?
Although any cell under approximately 40 µm can theoretically be sorted, practical limitations arise from cell health rather than cell size. FACS exposes cells to pressure and shear force, which can be stressful for fragile populations such as primary neurons or delicate epithelial cells.
Rare populations also present challenges. When collecting sufficient numbers of rare cells, sorting time may become prolonged, increasing the risk of apoptosis or stress-induced transcriptional changes. In such cases, pre-enrichment strategies or performing the workflow in smaller, freshly prepared batches can help preserve viability.
What is the required quality of single-cell suspensions for FACS?
The quality of the input cell suspension strongly affects sorting success. Suspensions containing debris, aggregates, or excessive dead cells increase the likelihood of clogging, complicate gating, and significantly prolong sorting time. These issues ultimately reduce viability and impact sequencing quality.
A high-quality single-cell suspension should have minimal debris, minimal clumping, and good pre-sort viability. Filtering through a 40-µm strainer immediately before sorting is one of the most effective ways to prevent clogging. If a sample tends to sediment or aggregate, gentle mixing during the sort helps maintain even cell distribution.
When samples contain a large number of red blood cells, RBC lysis can reduce event density and shorten the overall sorting duration, minimizing stress on the cells.
What should you consider when choosing fluorescent antibodies for FACS prior to single-cell sequencing?
Single-cell transcriptomic workflows require intact, viable cells. Therefore, staining should focus on surface markers, as intracellular staining requires fixation and permeabilization, which compromise RNA integrity.
Fluorophore selection should match both the anticipated antigen abundance and the sorter’s laser/detector configuration. Bright fluorophores such as PE work well for low-abundance markers, while FITC and other lower-intensity fluorophores are suitable for highly expressed antigens.
Viability dyes such as DAPI, PI, or 7-AAD are commonly used to exclude dead cells during sorting. When designing a panel, it is important to avoid spectral overlap between these dyes and the fluorophores used for surface markers. Reviewing the sorter’s channel layout in advance helps prevent spillover and ensures accurate gating.
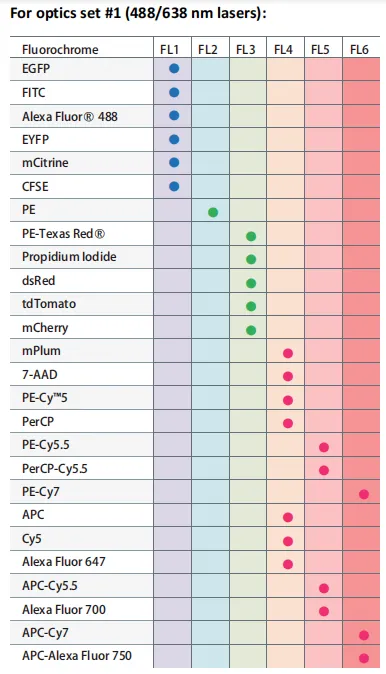
Figure 2. Fluorochrome detection channels for the SONY SH800S flow cytometer (488 nm and 638 nm laser configuration)
How should controls be set up for flow cytometry sorting?
Reliable gating depends on the proper use of controls. Unstained controls help establish baseline voltages and define negative thresholds. Biological controls, such as untreated or wild-type samples, provide reference expression patterns.
Compensation controls—using either single-stained cells or compensation beads—correct for spectral overlap between fluorophores. Compensation beads are often preferred because they generate strong, uniform signals and help preserve valuable biological samples.
For more complex staining panels, fluorescence-minus-one (FMO) controls can help define gating boundaries for dim markers or subsets with overlapping features.
How can viability loss be minimized during FACS?
Maintaining viability during sorting is crucial, as post-sort cell health directly influences single-cell sequencing metrics. Instrument calibration should be completed before sample preparation to minimize the time cells sit waiting.
If available, temperature-controlled chambers set to 5–10°C help protect cells by reducing metabolic activity. Selecting a larger nozzle and using lower sorting pressure also reduces mechanical stress.
On the sample-handling side, cells should be kept on ice, freshly dissociated, and filtered through a 40-µm strainer before sorting. Collection tubes should contain chilled buffer to cushion the impact as sorted cells land. For large experiments, dividing samples into smaller batches prevents any portion of the suspension from being subjected to prolonged stress.
Conclusion
Flow cytometry and FACS are powerful tools for preparing high-quality cells for single-cell sequencing. By optimizing nozzle size, suspension quality, staining strategies, and control design—and by reducing stress throughout the sorting process—researchers can significantly improve post-sort viability and downstream sequencing performance.
These considerations are especially important when working with fragile tissues, rare populations, or complex study designs where every viable cell matters.
Planning a single-cell Sequencing project?
With over eight years of dedicated experience in single-cell and spatial multi-omics, Omics Empower operates its own laboratories equipped with 10x Genomics and MobiDrop platforms, enabling reliable workflows for fresh, frozen, and FFPE samples.
Researchers worldwide trust our data: more than 400 peer-reviewed publications have been generated using our single-cell and spatial transcriptomics services, including studies in Nature, Science, and Cell. From library preparation to bioinformatics and publication-ready figures, we deliver end-to-end support to help you advance your next single-cell project.