Understanding how cells behave within their native tissue environment is critical for both basic biological research and translational studies. Each cell carries not only molecular information but also spatial context that can reveal insights into development, disease progression, and tissue structure.
To support this level of insight, 10x Genomics has developed Xenium In Situ, a next-generation spatial transcriptomics platform that offers single-cell and even subcellular resolution. Unlike conventional technologies that rely on in situ capture followed by sequencing, Xenium uses padlock probe-based rolling circle amplification (RCA) and fluorescence imaging to enable highly sensitive, specific, and multiplexed gene expression detection—all within intact tissue sections.
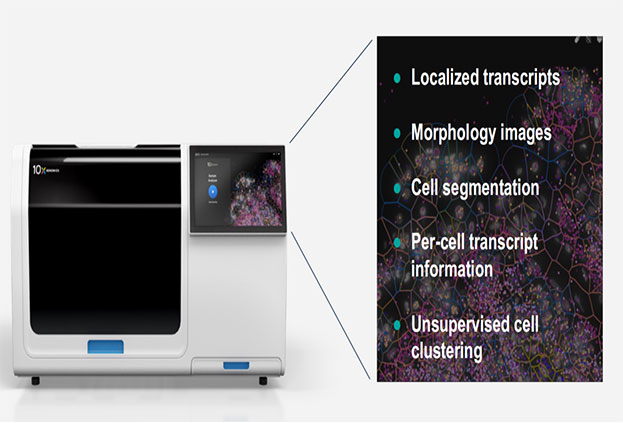
Figure 1. Overview of Xenium In Situ Technology
Xenium uses padlock probes, which feature two gene-specific binding arms and a central region designed to hybridize with fluorescent readout probes. Once both ends hybridize precisely to the target mRNA in situ, the probe forms a circular structure that undergoes rolling circle amplification. This process generates localized DNA concatemers rich in readout sequences.
These amplified products are sequentially hybridized with fluorescent probes and imaged, resulting in spatially encoded fluorescence barcodes that identify the gene of interest. Probe concentrations can be finely adjusted based on expected expression levels, optimizing both sensitivity and specificity for each target gene.
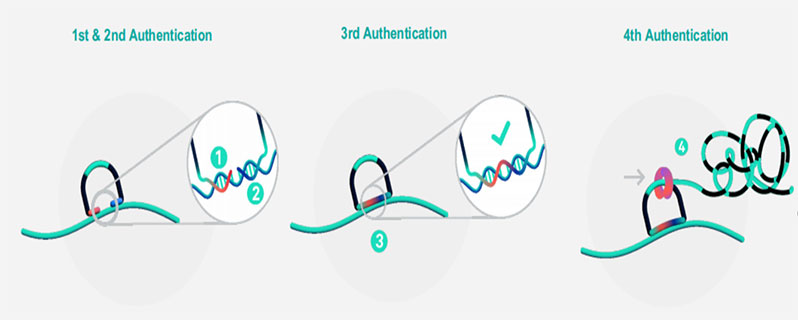
Figure 2. Workflow of Padlock Probe Hybridization and Rolling Circle Amplification
Tissue sections (fresh frozen or FFPE) are placed on Xenium slides. Fresh frozen samples are fixed and permeabilized; FFPE samples are dewaxed and undergo crosslink reversal.
Padlock probes hybridize to target mRNA and form circular structures upon correct binding. RCA generates gene-specific amplicons directly within tissue.
The sample is stained for cell segmentation (e.g., membrane and nuclear dyes), autofluorescence is quenched, and DAPI is used for nuclear labeling.
Fluorescent probes hybridize to the RCA products, and high-resolution imaging captures the spatial barcodes. The resulting images are decoded to determine gene identity and spatial location.
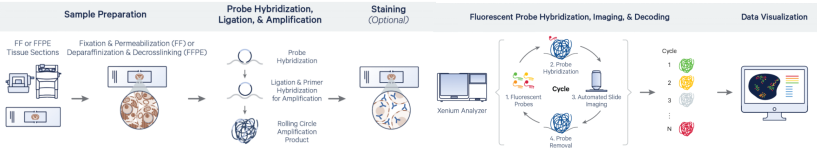
Figure 3. Xenium Workflow Overview
Unlike the Visium HD platform, which uses spatial barcoding and sequencing after probe capture, Xenium identifies transcripts via fluorescence-based in situ hybridization. Each gene is detected by a unique fluorescent barcode, enabling direct localization at the single-cell—or even subcellular—level.
Additionally, Xenium captures a much larger tissue area per slide, allowing for broader spatial coverage without compromising resolution.
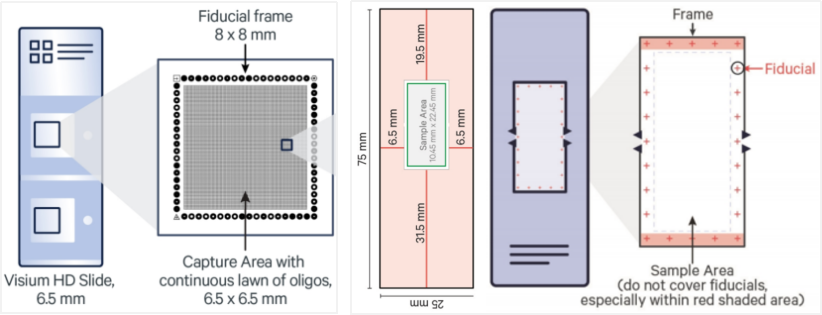
Figure 4. Comparison Between Xenium and Visium HD Slides
Xenium achieves high reproducibility across tissue types. For instance, adjacent sections tested with the Xenium 5000 panel show transcript correlation coefficients as high as 0.98.
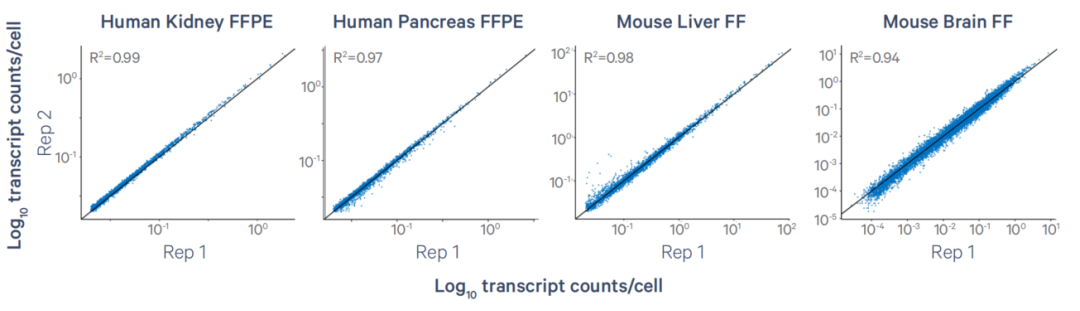
Figure 5. Xenium 5000 Panel Performance Across Species
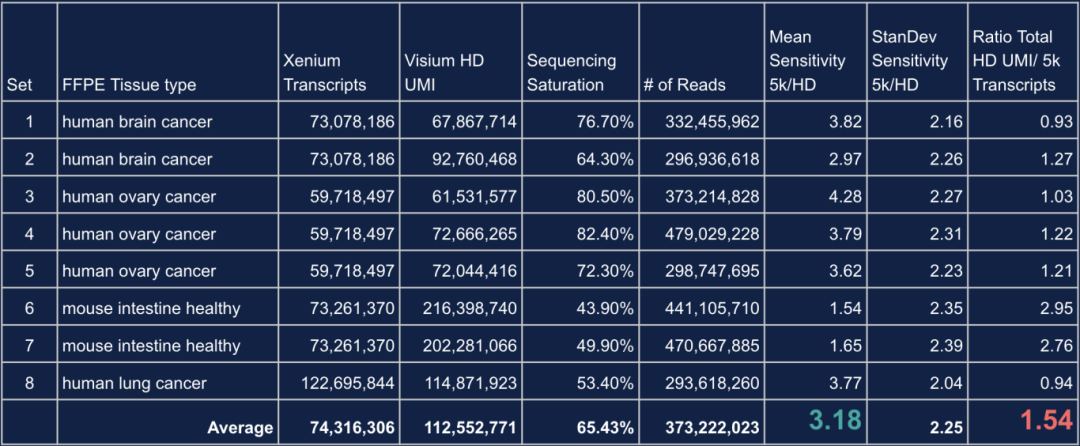
Its padlock-probe and RCA-based detection method offers high sensitivity, including detection of isoforms and SNPs. Compared to Visium HD, Xenium demonstrates:
3.18× higher single-gene sensitivity, depending on sequencing depth
Slightly lower total transcript yield (~1.54× less than Visium HD), highlighting complementary strengths between platforms
Table 1. Comparison of Performance Metrics Between Visium HD and Xenium
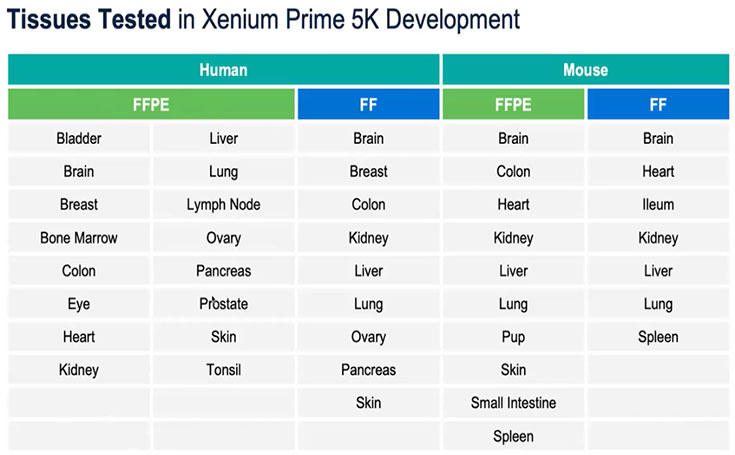
Xenium offers multiple panel options to meet diverse research needs:
A pre-designed 5000-gene panel for broad screening
The ability to customize up to 100 additional genes on top of the core panel
Fully custom panels up to 480 genes, available for almost any species
This flexibility supports both discovery-based and hypothesis-driven research.
Table 2. Xenium 5000 Panel Compatibility with Human and Mouse Tissues
Accurate cell segmentation is crucial for defining cell types and spatial organization. Xenium introduces multimodal segmentation, combining multiple fluorescent stains and machine learning to objectively define cell boundaries—even for irregularly shaped cells like fibroblasts.
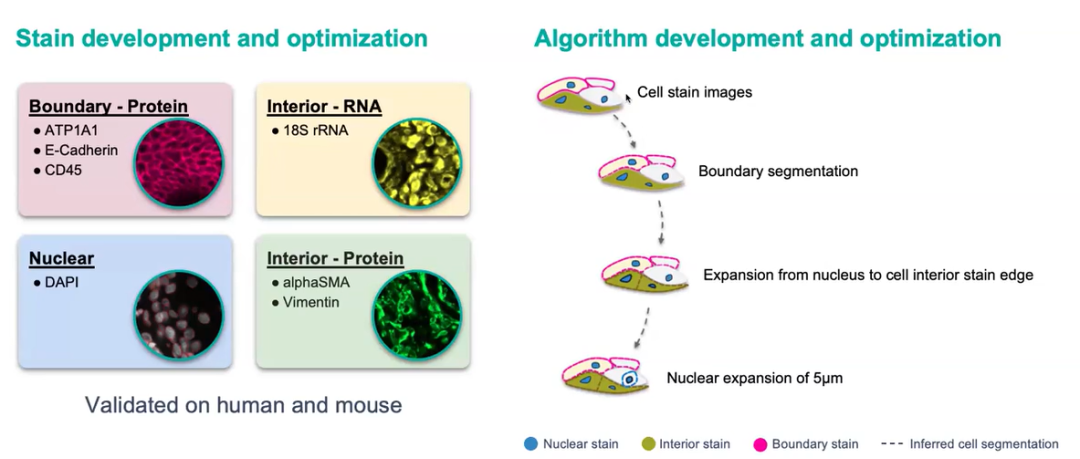
Figure 6. Multimodal Segmentation Workflow
This overcomes limitations of traditional nucleus-based segmentation, which assumes spherical cell morphology.
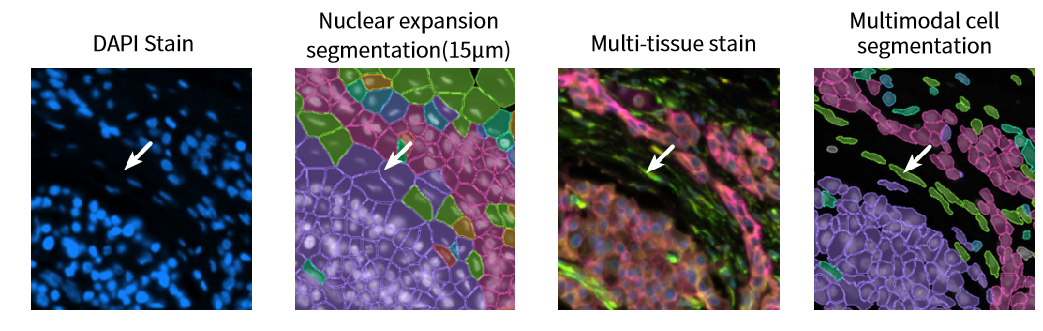
Figure 7. Improved Accuracy in Irregular Cell Morphologies
Xenium’s non-destructive assay preserves tissue morphology, allowing researchers to perform follow-up assays (e.g., immunofluorescence or histology) on the same FF or FFPE section. This opens the door to true multi-omic analysis from a single slide.
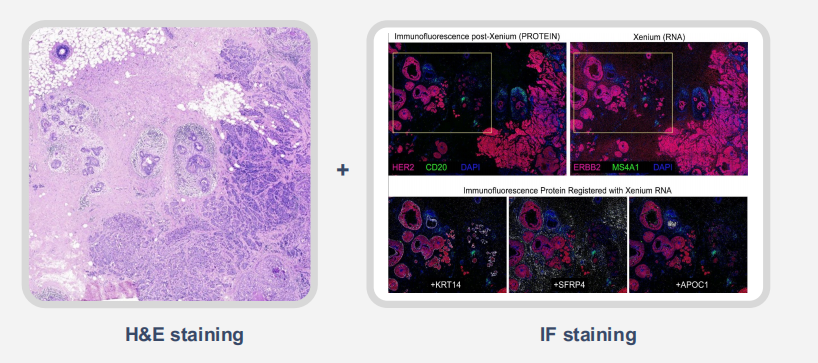
Figure 8. Post-Xenium Compatibility with Additional Assays
Xenium includes powerful onboard software (Xenium Onboard Analysis, XOA) that processes and decodes data in real time during imaging.
For downstream analysis, Xenium Explorer offers intuitive visualization of:
Tissue morphology
Cell segmentation and classification
Transcript density and spatial distribution
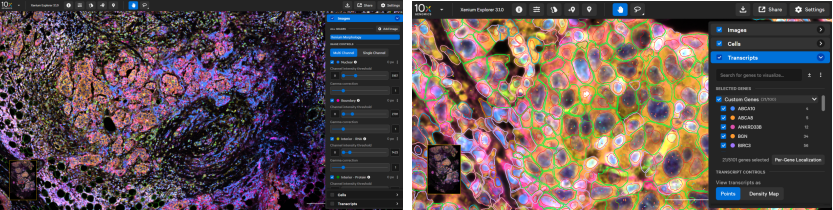
Figure 9. Xenium Explorer Output on FFPE Breast Cancer Sample
Compared to traditional in situ hybridization techniques, Xenium offers rapid, high-throughput spatial profiling of candidate genes at both the single-cell and subcellular levels. Its advanced multimodal cell segmentation algorithm enables more accurate delineation of cells with irregular shapes or varying sizes, a common challenge in complex tissue environments.
In such scenarios, Xenium outperforms earlier platforms like Visium V1, V2, and HD, making it particularly suitable for studies that demand high resolution and spatial precision.
Xenium also integrates seamlessly with single-cell RNA-seq and other spatial transcriptomics platforms. A common workflow involves using whole-transcriptome profiling to identify potential biomarkers, followed by targeted, spatially resolved validation using Xenium panels. This two-step approach has become a standard strategy for spatial transcriptomic validation and biomarker discovery.
At Omics Empower, we specialize in integrating Xenium with single-cell RNA sequencing and other spatial transcriptomics platforms. With extensive project experience, we support researchers across the entire workflow—from sample preparation to bioinformatics analysis.
If you're planning a spatial biology project, feel free to schedule a meeting with our scientific team. We're here to help you make the most of Xenium and design efficient, high-impact experiments.
*Credit: All figures and data presented in this article are credited to 10x Genomics unless otherwise noted.
Singapore Global Headquarters: 112 ROBINSON ROAD #03-01
Germany: Kreuzstr. 60, 40210 Düsseldorf
United States: 2 Goddard, Irvine, CA 92618
Hong Kong: Flat 1019B, 10/F, Liven House, No. 61–63 King Yip Street, Kwun Tong
Singapore Global Headquarters: 112 ROBINSON ROAD #03-01
Germany: Kreuzstr. 60, 40210 Düsseldorf
United States: 2 Goddard, Irvine, CA 92618
Hong Kong: Flat 1019B, 10/F, Liven House, No. 61–63 King Yip Street, Kwun Tong