As spatial biology continues to reshape life science research, selecting the right spatial transcriptomics platform has become a key decision. Whether you’re analyzing FFPE samples, prioritizing subcellular resolution, or exploring high-throughput options, today's commercial solutions—from 10x Genomics to BGI —offer diverse technologies tailored to different experimental needs. In this article, we break down the strengths and technical highlights of leading platforms to help you make the best choice for your research.
Robust and Widely Used for Fresh-Frozen Tissue
Overview:
Visium 1.0, launched in 2019, integrates barcoded capture probes into slide-based spatial arrays. Each slide contains four 6.5 × 6.5 mm capture areas, each with 5,000 barcoded spots (55 μm diameter, 100 μm spacing), making it ideal for broad tissue compatibility and morphological validation.
Strengths:
Supports polyA+ RNA from animals and plants
Compatible with H&E staining on the same section
Works with fresh-frozen and -80°C preserved samples
Proven across multiple species: human, mouse, zebrafish, Arabidopsis, etc.
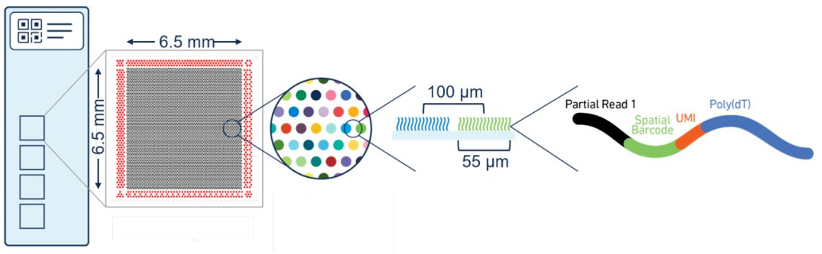
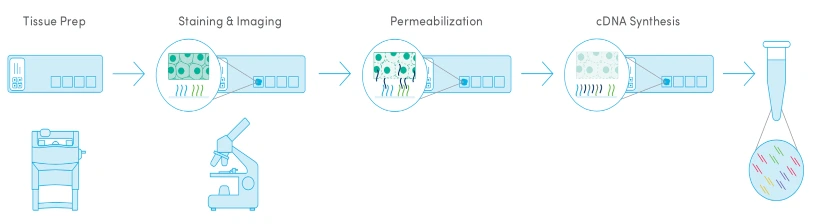
Best for:
General-purpose spatial transcriptomics with moderate resolution and flexible tissue input.
Optimized for FFPE Tissues with Larger Capture Area
Overview:
Visium 2.0 (CytAssist FFPE) builds on Visium 1.0 with redesigned probes that allow FFPE compatibility without requiring polyA tails. A new 11 × 11 mm chip increases spot count to ~14,000 while maintaining the same resolution.
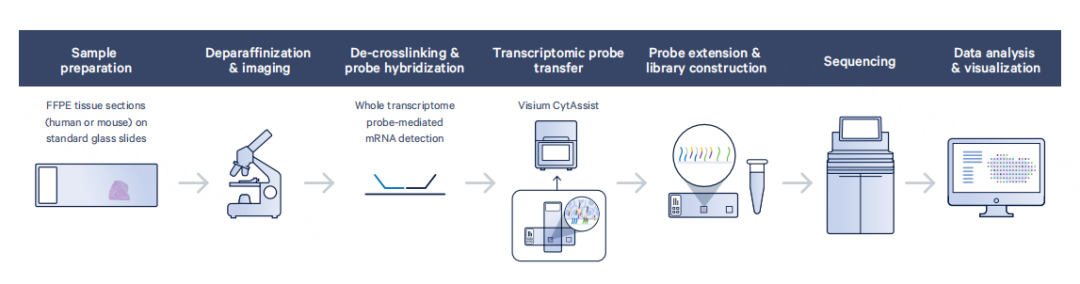
Strengths:
Enables spatial profiling of archival FFPE samples
No permeabilization required—streamlined workflow
H&E stain compatibility retained
Compatible with FFPE, fresh, and frozen samples
Available for human and mouse tissues
Best for:
Researchers working with rare, clinical, or archived FFPE specimens.
High-Resolution Platform with Submicron Spot Spacing
Overview:
Stereo-seq offers spatial transcriptomics with exceptional resolution using dense spot arrays (220 nm diameter, 500 nm spacing). Launched commercially in 2022, it supports both 10×10 mm and 5×5 mm capture chips using poly-dT RNA capture.
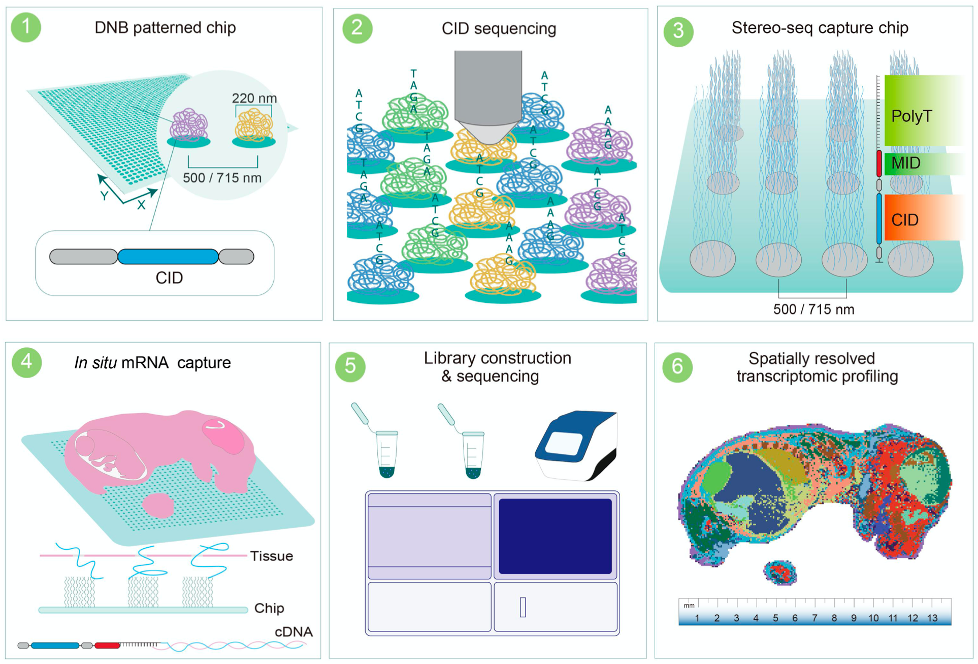
Strengths:
Ultra-high spatial resolution—approaching single-cell level
Broad species and sample compatibility (fresh and -80°C)
Adjustable resolution for tailored analysis
Proven in human, mouse, zebrafish, Arabidopsis, and more
Best for:
Researchers prioritizing fine spatial mapping, single-cell-level resolution, and scalable output.
Targeted Spatial Profiling with Single-Cell Precision
Overview:
Xenium offers high-resolution in situ gene detection via padlock probe technology. Its chip size is 12 × 24 mm, with panels supporting detection of ~300 genes (customizable up to 500). Unlike sequencing-based tools, Xenium provides direct spatial imaging at subcellular resolution.
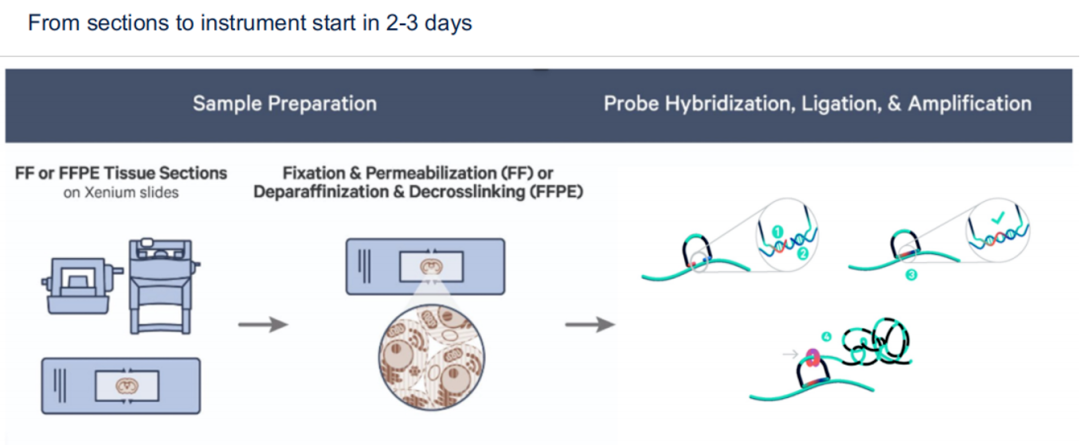
Strengths:
Subcellular-level spatial resolution
Compatible with FFPE, fresh, and frozen tissues
Non-destructive: post-assay H&E and IHC still possible
Targeted panels for cancer and developmental research
Designed for human and mouse tissues
Best for:
Users seeking targeted gene panels with ultra-high-resolution imaging of spatial gene expression.
There is no one-size-fits-all solution in spatial transcriptomics. Each platform offers unique strengths depending on your research priorities:
Research Priority | Best Platform |
FFPE sample compatibility | Visium 2.0 (CytAssist FFPE), Xenium |
Highest spatial resolution | Stereo-seq, Xenium |
Broad gene detection (whole-transcriptome) | Visium 1.0/2.0, Stereo-seq |
Targeted, single-cell-level detection | Xenium |
Broad species/sample coverage | Visium 1.0, Stereo-seq |
With spatial omics still in its growth phase, now is the time to explore new technologies and match the right platform to your experimental goals.
At Omics Empower, we offer full-service spatial transcriptomics solutions based on industry-leading platforms—including 10x Genomics Visium, Xenium, and BGI Stereo-seq.
Whether you're working with FFPE or fresh-frozen tissue, human or plant samples, our team provides end-to-end support—from sample processing and library preparation to bioinformatics analysis. Our services are designed to meet diverse research needs in oncology, neuroscience, developmental biology, and beyond.
Supporting Global Research
With lab operations in the U.S., Europe, and Asia, we provide responsive support for international projects and multi-site studies. Coordinated logistics ensure timely delivery and sample stability—no matter where your research takes place. Contact us today for a consultation!
Credit: Equipment illustrations and platform schematics featured in this article are sourced from official materials provided by © 10x Genomics and © BGI Genomics.
Singapore Global Headquarters: 112 ROBINSON ROAD #03-01
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: 2 Goddard, Irvine, CA 92618
Hong Kong: Room 618, Building 6, Phase One, Hong Kong Science Park, No. 6 Science Park West Avenue, Pak Shek Kok, New Territories, Hong Kong
Singapore Global Headquarters: 112 ROBINSON ROAD #03-01
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: 2 Goddard, Irvine, CA 92618
Hong Kong: Room 618, Building 6, Phase One, Hong Kong Science Park, No. 6 Science Park West Avenue, Pak Shek Kok, New Territories, Hong Kong