10x Genomics released Visium HD in January 2024—a next-generation spatial transcriptomics platform capable of mapping gene expression at near single-cell resolution. Designed for FFPE (formalin-fixed, paraffin-embedded) tissues, this platform has gained considerable attention from the biomedical research community.
At Omics Empower, we’ve compiled answers to the most frequently asked technical questions researchers have raised when planning Visium HD projects. This guide serves as an entry point for understanding the platform’s unique features, workflow, sample requirements, and data outputs.
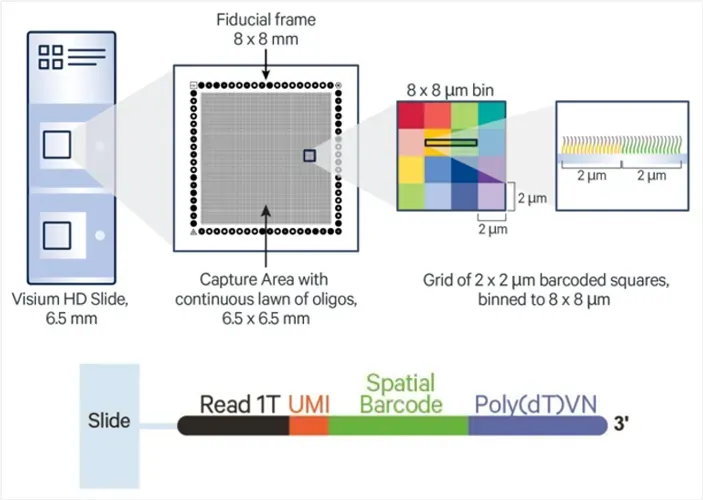
Visium HD introduces a redesigned capture slide, offering subcellular resolution and high spatial fidelity:
Each slide contains two 6.5 × 6.5 mm capture areas.
The capture areas are densely packed with 2 × 2 µm barcoded oligo squares.
Suggested analysis bin size is 8 × 8 µm, enabling near single-cell spatial resolution.
Each oligo contains an Illumina Read 1T sequence, UMI, spatial barcode, and poly(dT) tail for capturing polyadenylated RNA.
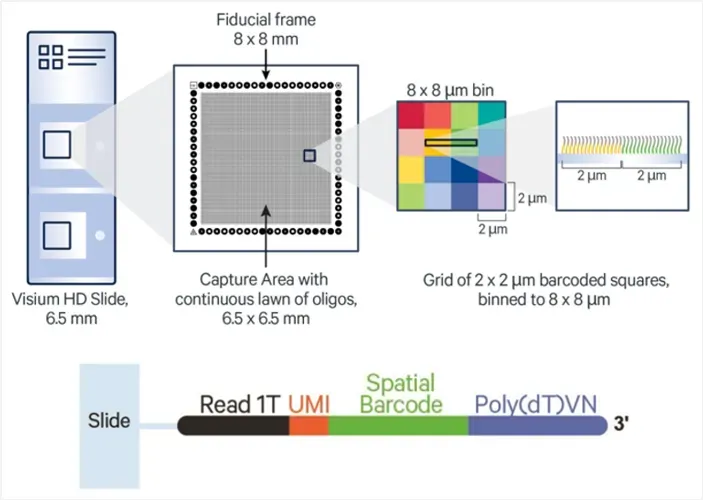
Figure 1. Schematic of Visium HD Slide Capture Area (top) and Oligonucleotide Structure (bottom)
Visium HD shares the same core technical principles as Visium CytAssist (V2), which include:
(1) Targeted probes based on annotated genes in human and mouse genomes.
Each probe consists of a left-hand side (LHS) and a right-hand side (RHS):LHS contains part of the Read 2S sequence and a 25 bp sequence complementary to the left side of the target mRNA.
RHS contains a 25 bp sequence complementary to the right side of the mRNA and a 30 bp poly(A) tail.
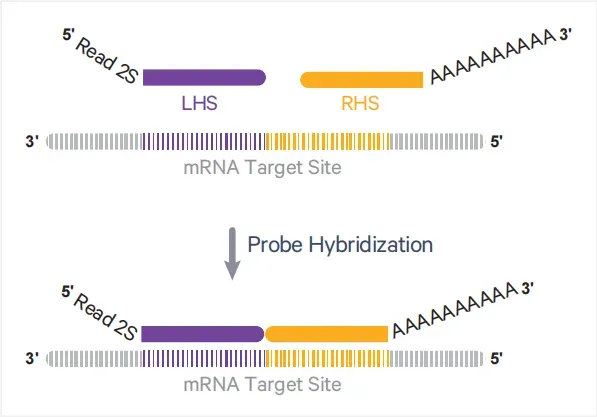
Figure 2. Schematic of Probe Structure
(2) CytAssist-mediated in situ transfer.
During the transfer process, the tissue slide with hybridized probes is tightly aligned with the Visium HD slide using the CytAssist instrument. RNase treatment digests the tissue RNA, releasing probe-mRNA hybrids. The poly(A) tail of the probe hybridizes with the poly(dT) oligos on the Visium slide, enabling spatial capture.
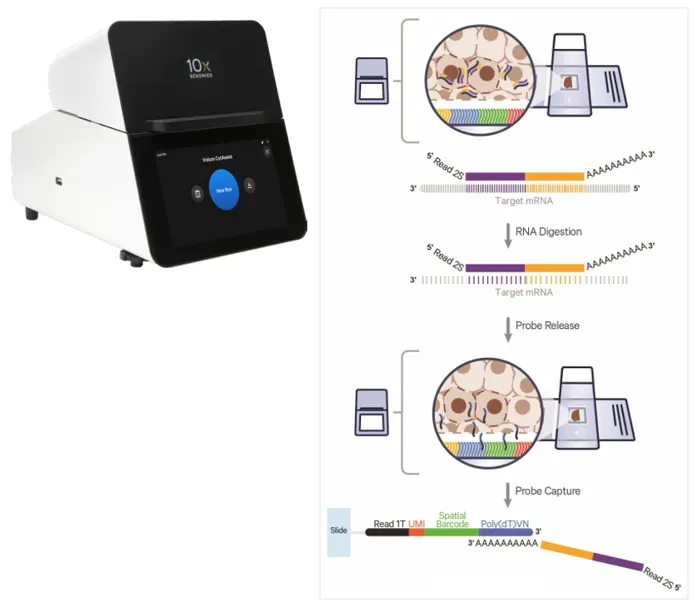
Figure 3. Principle of Tissue Transfer via CytAssist
Visium HD currently supports only FFPE samples from human and mouse tissues. The sample preparation protocol is identical to that of Visium CytAssist (V2). For high-quality results, tissue sections must exhibit well-preserved nuclear morphology, good RNA integrity, and no detachment during processing.Key considerations include:
(1) Paraffin blocks are preferred.
(2) If only FFPE slides are available, ensure:Section thickness is 5 µm with proper drying.
Use 10x-recommended slides, and follow the specific placement guidelines for each slide model
For tissues prone to detachment (e.g., skin, blood vessels, fat, tendon, breast, lung, colon, multinucleated or connective tissues), adhesion slides such as Schott H-3D (PN-1800434) are strongly recommended.
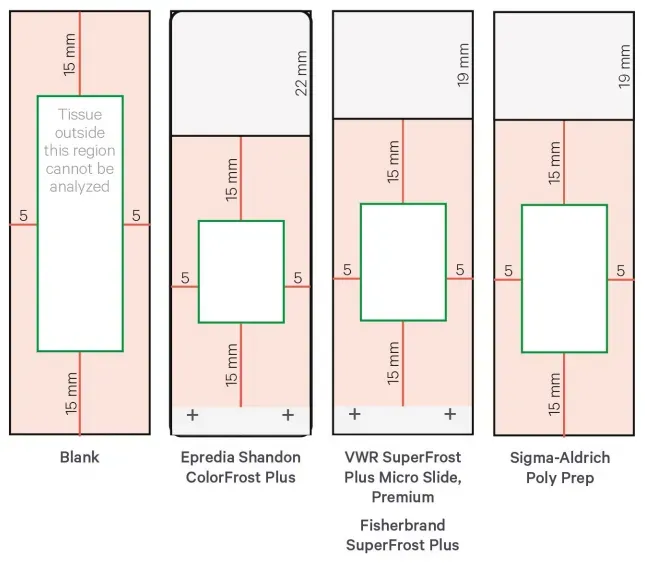
Figure 4. Recommended Slide Types and Tissue Placement Areas by 10x Genomics (indicated in green)
(3) Transport conditions: 4°C. Slides should remain dry throughout shipping (refer to Omics Empower’s sample submission guide).
(4) RNA quality requirement: DV200 > 30%, with a normal RNA profile.
The Visium HD workflow is identical to Visium CytAssist (V2) and includes the following steps:
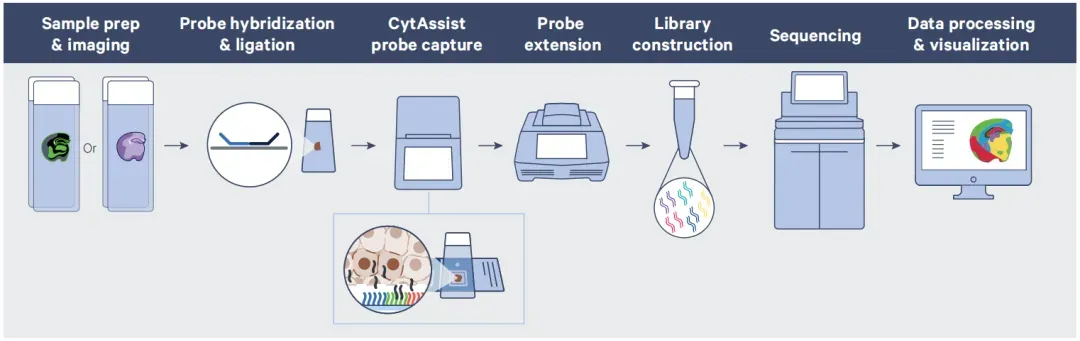
Figure 5. Visium HD Workflow Overview
(1) Sample preparation and imaging: FFPE tissue is sectioned onto the effective capture area of a 10x-recommended slide. This is followed by deparaffinization, H&E staining, image scanning, destaining, and decrosslinking. Probe hybridization is performed immediately afterward.
(2) After overnight hybridization and washing, the probes are ligated.
(3) The CytAssist instrument performs the slide transfer.
(4) Probes are extended on the Visium HD slide using a thermal cycler.
(5) Captured probes are eluted and undergo pre-amplification and library construction.
(6) Libraries are sequenced and analyzed.
The Visium HD library structure is as follows:
Read 1 contains the spatial barcode and UMI (total length: 43 bp)
Read 2 contains the 50 bp probe insert sequence
Sequencing is performed in PE150 mode, with ~275 million reads recommended per capture area.
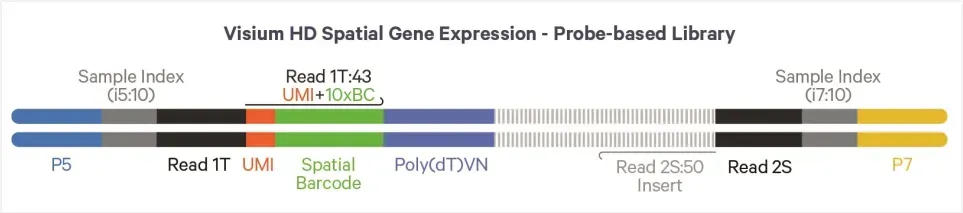
Figure 6. Structure of the Visium HD Probe Library
Since Visium HD is used for FFPE samples, which require water spreading during sectioning, tiling multiple samples increases the risk of detachment—especially for fragile tissues. For this reason, tiling is generally discouraged. If necessary, no more than two tissue pieces should be placed within the same capture area.
For human samples, Visium HD uses the same V2 probe design as Visium CytAssist (V2), with three probe pairs per gene.
For mouse samples, Visium HD uses an upgraded version. While CytAssist (V2) uses only one probe pair per gene (V1), Visium HD uses three probe pairs per gene (V2). This provides improved detection efficiency and a higher median gene count.
10x Genomics has published a list of FFPE tissue types validated for Visium HD. In our experience, FFPE samples that have worked successfully on Visium CytAssist generally perform well on Visium HD as well. Conversely, samples that previously failed with CytAssist are unlikely to be successful on Visium HD. For assistance with specific sample types, please feel free to contact our technical specialists.
Omics Empower is a 10x Genomics Certified Service Provider. If you are interested in how our services can support your research, please reach out to our team. Whether you need assistance with sample preparation, single cell sequencing, spatial transcriptomics sequencing, or data analysis, we can handle every step of the process to ensure high-quality results. Contact us today for a consultation!
**Credit: All figures and data presented in this article are credited to 10x Genomics unless otherwise noted.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong