
A recent study from Dr. Jiulin Du’s research group has uncovered a novel mechanism linking neural activity to the development of the brain’s lymphatic system—mediated by a specific population of glial cells. The work, published in Cell on April 30, 2025, is titled “Neural-activity-regulated and glia-mediated control of brain lymphatic development.”
Using single-cell RNA sequencing, the team identified a subpopulation of radial astrocyte-like glia marked by slc6a11b expression. These cells were found to secrete Vegfc, a key factor that drives the development of meningeal lymphatic endothelial cells (muLECs). Interestingly, the secretion of Vegfc is modulated by neural activity, suggesting a previously unrecognized crosstalk between neurons and the developing lymphatic system.
To further dissect this mechanism, the study employed conditional gene editing and in vivo calcium imaging, revealing that the radial astrocytes not only respond to neural cues but actively regulate lymphatic vessel formation during brain development. This discovery opens new therapeutic avenues for neurological disorders associated with lymphatic dysfunction.
*Service Provider: Omics Empower provided Single-cell transcriptome sequencing services.If you're looking for similar single-cell or spatial solutions support in your own research, our expert would be happy to help.
The nervous system can regulate immune responses through defined anatomical pathways under both physiological and pathological conditions. However, whether it also plays a role in shaping immune system development—and how—has remained unclear.
Previous research has shown that the development of meningeal lymphatic endothelial cells (muLECs) depends on Vegfc–Vegfr3 signaling. muLECs help clear metabolic waste from the brain and are essential for maintaining immune and fluid homeostasis. Thus, identifying the cellular source of Vegfc and understanding whether neural activity affects muLEC development are key to improving therapeutic strategies for brain lymphatic dysfunction.
This study demonstrates that neural activity regulates the development of meningeal lymphatic endothelial cells (muLECs) in zebrafish. Through neural modulation experiments, the researchers showed that changes in brain activity directly affect muLEC formation.
Using single-cell RNA sequencing, they identified slc6a11b⁺ radial glia-like cells (RAs) as the primary source of Vegfc, a key signaling molecule driving muLEC development. The team also used conditional gene editing and in vivo calcium imaging to investigate the functional role of RAs and found that these cells are in direct contact with muLECs.
Furthermore, by combining calcium imaging, eye removal, and pharmacological inhibition experiments, the study confirmed that both RA activity and Vegfc expression are closely linked to neural activity. These results indicate that neural signals regulate muLEC development by controlling Vegfc production in RAs.
To investigate the developmental dynamics of meningeal lymphatic endothelial cells (muLECs), the researchers generated transgenic zebrafish lines. They observed that muLECs formed a ring-like structure along the boundary between the soma and fiber layers of the optic tectum (OT)—a key region involved in visual processing—and gradually expanded to cover the surface of the OT (Fig. 1A–B).
When neural activity was pharmacologically suppressed using tricaine, ring formation was blocked, and the number of muLECs on the OT surface was significantly reduced (Fig. 1C–G).
To assess the role of sensory input, the researchers removed the eyes of the zebrafish larvae to reduce visual stimulation. This also led to a marked reduction in muLEC numbers on the OT surface (Fig. 1H–P).
In contrast, when electrical stimulation (ES) was applied to enhance neural activity, the number of muLECs increased significantly in the stimulated larvae (Fig. 1Q–R).
Together, these results provide strong evidence that neural activity plays a regulatory role in the development of brain lymphatic structures.
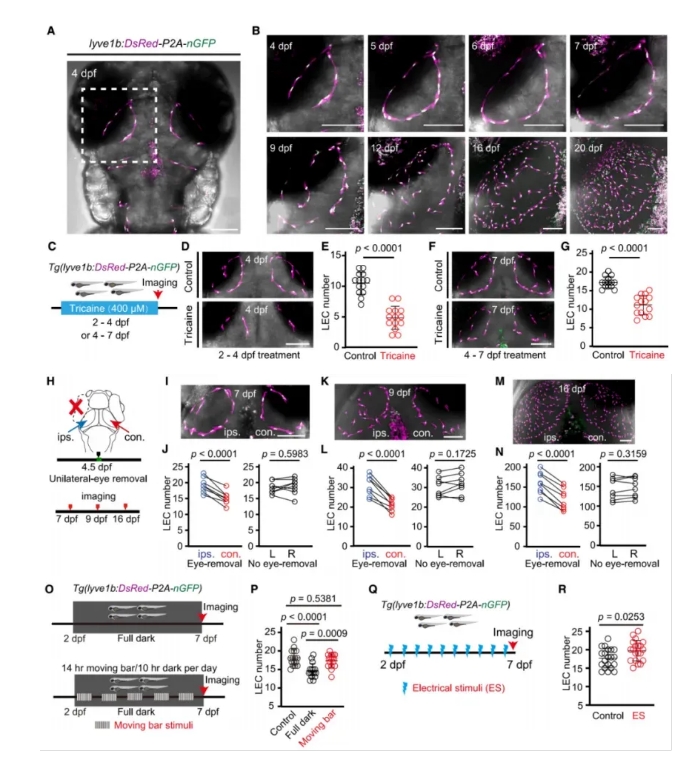
Figure 1. Neural Activity Regulates muLEC Development in Zebrafish
Vegfc and Vegfd are secreted proteins known to play essential roles in lymphatic vessel development in vertebrates. To identify the source of these factors in the brain, the authors performed single-cell RNA sequencing on whole-brain samples from zebrafish at three developmental stages.
They found that radial glia-like cells (RAs) were the predominant cell type expressing both Vegfc and Vegfd (Fig. 2A–B).
Using a knock-in (KI) reporter zebrafish line, the researchers further visualized Vegfc⁺ RAs, which were mainly distributed throughout the brain. These cells extended processes toward the choroid vascular plexus (CVP) and reached the brain surface, where they made direct contact with muLECs (Fig. 2C–E).
Subtype analysis of RAs at the single-cell level, combined with in situ hybridization, confirmed that slc6a11b⁺ RAs were the primary source of Vegfc and Vegfd in the brain. These cells exhibited the same morphology as Vegfc⁺ RAs and maintained direct contact with muLECs (Fig. 2F–K).
Collectively, these findings indicate that slc6a11b⁺ RAs regulate muLEC development through Vegfc signaling.
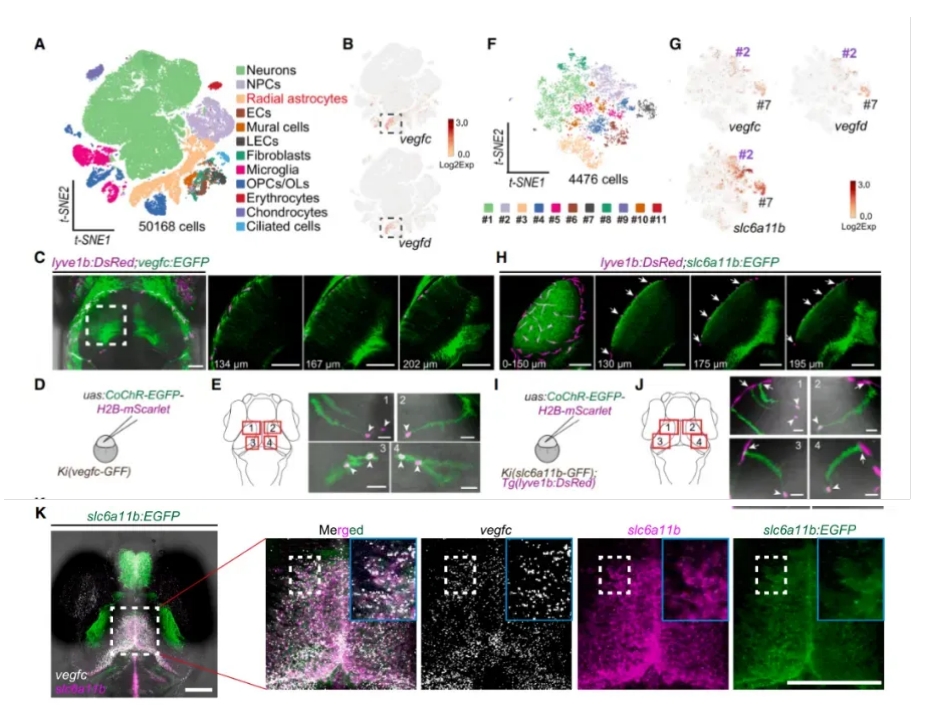
Figure 2. slc6a11b⁺ RAs Are the Primary Source of Vegfc/Vegfd in the Brain
To further explore whether slc6a11b⁺ radial glia-like cells (RAs) regulate muLEC development, the authors conducted functional loss-of-function experiments.
They treated zebrafish with metronidazole (MTZ) to selectively ablate slc6a11b⁺ RAs. This led to impaired muLEC development, as evidenced by the failure of ring structure formation and a significant reduction in muLEC numbers (Fig. 3A–C).
Next, blocking the Vegfc–Vegfr3 signaling pathway disrupted muLEC formation. Conversely, overexpressing Vegfc specifically in slc6a11b⁺ RAs enhanced muLEC development (Fig. 3D–E).
To test cell-type specificity, the researchers performed conditional Vegfc knockout and overexpression in both slc6a11b⁺ RAs and pdgfra⁺ fibroblasts. They found that only manipulation in slc6a11b⁺ RAs had a significant impact, confirming that these cells play a dominant role in regulating muLEC development (Fig. 3F–J).
Together, these results demonstrate that slc6a11b⁺ RAs are essential for muLEC development through the Vegfc–Vegfr3 signaling axis.
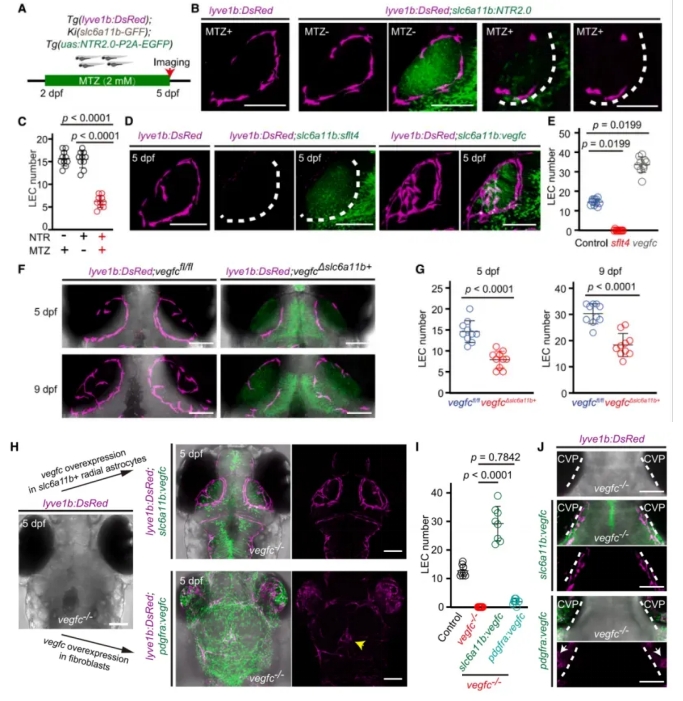
Figure 3. Vegfc Expression by slc6a11b⁺ RAs Is Critical for muLEC Development
To determine whether slc6a11b⁺ RAs and their expression of Vegfc are regulated by neural activity, the researchers conducted a series of functional imaging and perturbation experiments:
Using dual-color calcium imaging, they found that slc6a11b⁺ RAs responded to light stimulation with increased calcium activity, similar to neurons in the optic tectum (OT) neuropil (Fig. 4A–D).
In unilateral eye removal experiments, spontaneous calcium activity in contralateral OT RAs was impaired compared to the ipsilateral side (Fig. 4E–H).
Vegfc expression in RA processes was significantly reduced on the side corresponding to the removed eye, although the number of slc6a11b⁺ RAs remained unchanged (Fig. 4I–L).
Blocking neural activity pharmacologically also resulted in reduced Vegfc expression in these cells (Fig. 4M–Q).
These results demonstrate that neural activity positively regulates Vegfc expression in slc6a11b⁺ RAs, and that reduced neural input leads to downregulation of Vegfc, thereby impairing muLEC development.
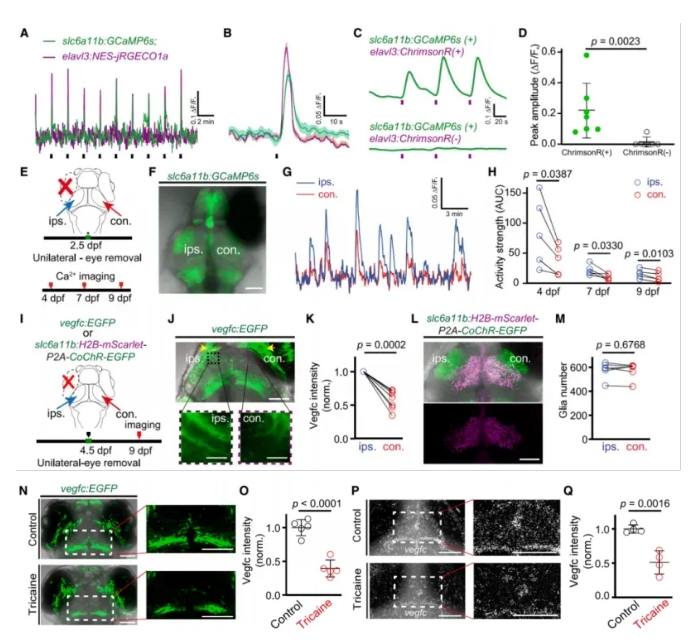
Figure 4. Activity and Vegfc Expression in slc6a11b⁺ RAs Are Coupled to Neural Signals
The researchers next examined whether neural activity influences muLEC development specifically through Vegfc expression in slc6a11b⁺ RAs.
They conducted neural activity perturbation experiments—including unilateral eye removal and moving visual stimuli—in zebrafish larvae with conditional deletion of Vegfc in slc6a11b⁺ RAs (Vegfc cKO^slc6a11b⁺^). In contrast to wild-type controls (Vegfc^fl/fl^), these manipulations had no effect on muLEC numbers in the Vegfc-deficient larvae (Fig. 5).
However, overexpression of Vegfc in slc6a11b⁺ RAs successfully rescued the loss of muLECs caused by visual deprivation.
These findings indicate that neural activity regulates muLEC development via glia-derived Vegfc, and that slc6a11b⁺ RAs act as a functional bridge between neural signals and lymphatic formation.
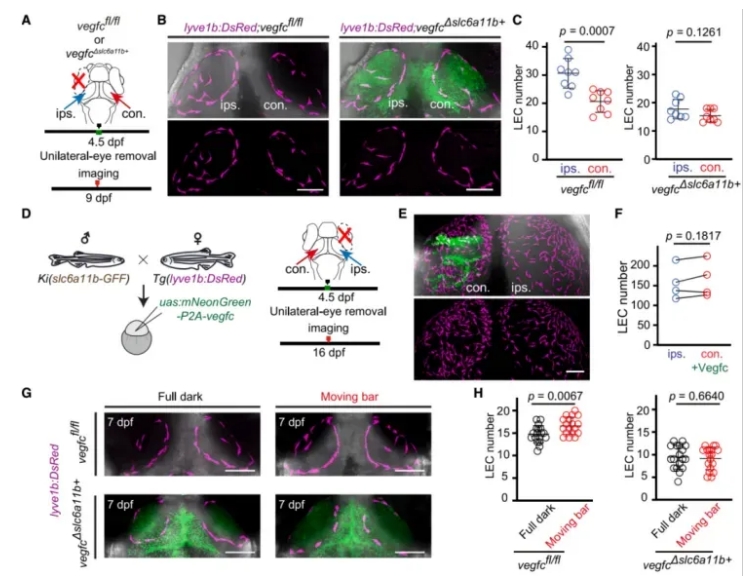
Figure 5. Neural Activity Regulates muLEC Development Through Vegfc Expression in slc6a11b⁺ RAs
The previous results demonstrated that slc6a11b⁺ radial glia (RAs) promote muLEC development through Vegfc expression in the brain. However, while RAs are located deep in the brain parenchyma, muLECs are confined to the meningeal surface, raising the question: What limits muLECs to the brain surface despite Vegfc being produced internally?
To investigate this, the researchers generated ccbe1 knockout zebrafish, and found that overexpressing Vegfc in slc6a11b⁺ RAs could not rescue the developmental defects in muLECs caused by ccbe1 loss (Fig. 6A–C).
Using a knock-in zebrafish line, they discovered that ccbe1 is expressed in fibroblasts localized to the brain surface (Fig. 6D). Furthermore, when the mature form of Vegfc was ectopically expressed in slc6a11b⁺ RAs, lymphatic endothelial cells (LECs) aberrantly grew inside the brain parenchyma, forming ectopic, lumenized vessels (Fig. 6E–G).
These findings suggest that the spatial precision of muLEC formation is controlled by two spatially distinct cell populations:
slc6a11b⁺ RAs within the brain parenchyma, which generate Vegfc
ccbe1⁺ fibroblasts at the brain surface, which enable Vegfc maturation and restrict its functional range
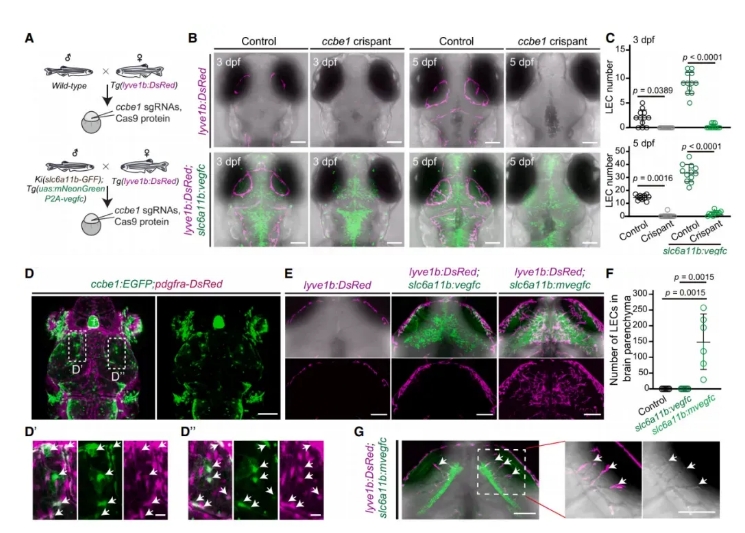
Figure 6. ccbe1⁺ Fibroblasts and slc6a11b⁺ RAs Cooperatively Regulate muLEC Patterning
slc6a11b⁺ radial glia (RAs) transport pro-Vegfc from within the brain to the meningeal surface. There, ccbe1⁺ fibroblasts cooperate with RAs to convert pro-Vegfc into its mature, active form at the interface between the brain parenchyma and meningeal fibroblasts.
This two-step, spatially coordinated mechanism ensures that Vegfc activity—and thus muLEC formation—is confined to the brain surface, preventing ectopic lymphatic development within the brain.
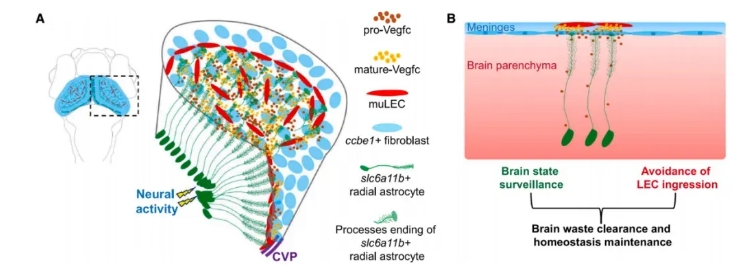
Figure 7. Working Model of Brain-Controlled muLEC Development
1. slc6a11b⁺ radial glia (RAs) are the primary source of Vegfc in the brain.
2. Vegfc produced by slc6a11b⁺ RAs is essential for muLEC development.
3. Neural activity regulates muLEC development by modulating Vegfc expression in slc6a11b⁺ RAs.
4. slc6a11b⁺ RAs and ccbe1⁺ fibroblasts work together to confine muLEC formation to the brain surface.
Li J, Liu MJ, Du WJ, Peng XL, Deng H, Zi HX, Shang HB, Du JL. Neural-activity-regulated and glia-mediated control of brain lymphatic development. Cell. 2025 Apr 22:S0092-8674(25)00410-6. doi: 10.1016/j.cell.2025.04.008.
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong
Germany: Arnold-Graffi-Haus / D85 Robert-Rössle-Straße 10 13125 Berlin
United States: (CA) 2 Goddard, Irvine, CA 92618
United States: (IL) 8255 Lemont Rd, #1, Darien, IL 60561
Hong Kong: Room 618, Building 6, Hong Kong Science Park, Pak Shek Kok, Hong Kong